2020 Volume 57 Issue 3 Pages 210-222
2020 Volume 57 Issue 3 Pages 210-222
Few studies have focused on the regulation of cytokine and avian β-defensin (AvBDs) expression for promoting immune defense in the avian intestine. The aim of this study was to investigate the effects of different Toll-like receptor (TLR) ligands (bacterial patterns) on the expression of proinflammatory cytokines (IL-1β and IL-6) and AvBDs (AvBD1, AvBD4, and AvBD7) in the chick intestine. The ileum and cecum of 3-day-old chicks were collected and examined histologically to identity the cells present in the intestinal mucosa. Other tissues were cultured with or without the TLR2, TLR4, and TLR21 ligands—Pam3CSK4, LPS, and CpG-ODN—for 1 or 3 h. The gene expression profiles of proinflammatory cytokines and AvBDs were determined in these tissues using real-time polymerase chain reaction (PCR). The mucosa of the ileum and cecum contained leukocytes, luminal and crypt epithelial cells, and other enterocytes. Pam3CSK4 tended to downregulate the expression of IL-1β, AvBD1, and AvBD7 in the ileum but upregulated their expression in the cecum. LPS downregulated the expression of IL-1β and IL-6 in both the ileum and the cecum, whereas it upregulated the expression of AvBD1, AvBD4, and AvBD7 in the cecum. CpG-ODN upregulated the expression of IL-6 and AvBD7 in the ileum and IL-1β in the cecum, and downregulated the expression of IL-1β and AvBDs in the ileum. We suggested that the expression levels of proinflammatory cytokines and AvBDs in the chick intestine are affected by TLR2, TLR4, and TLR21 ligands. Thus, these innate immune factors may be modulated by the luminal microbe complex in the intestine.
Colonization by pathogenic microorganisms, such as Salmonella and Campylobacter, may impair the health of chicks and cause bacterial contamination of poultry products. Gut-associated lymphoid tissues undergo development during the first few weeks of life (Bar-Shira et al., 2003). The innate immune system and maternal antibodies transported from the yolk play important roles for the protection of newly-hatched chicks against pathogenic microbial infection (Smith and Beal et al., 2008, Kovacs-Nolan et al., 2012). Therefore, innate immune function in the intestinal mucosa of chicks is important for defense against infection by microorganisms.
Toll-like receptors (TLRs) are pattern recognition receptors that recognize microbe-associated molecular patterns (MAMPs). In chickens, expression of TLR1-5, TLR7, TLR15, and TLR21 was observed in different intestinal segments in 2-day-old chicks (MacKinnon et al., 2009). TLR2, 4, and 21 are responsible for recognizing bacterial patterns. For example, TLR2 recognizes peptidoglycan and lipoproteins from Gram-positive bacteria (Keestra et al., 2007), whereas TLR4 recognizes lipopolysaccharide (LPS) from Gram-negative bacteria (Paul et al., 2013). TLR21 recognizes unmethylated CpG-oligodeoxynucleotides (ODN) from microorganisms (Keestra et al., 2010). Taha-abdelaziz et al. (2016) reported that in mononuclear cells isolated from chicken cecal tonsils, IL-1β expression increased after stimulation with Pam3CSK4 (a TLR2 ligand), LPS, and CpGODN, while LPS and CpG-ODN stimulation increased IL-6 expression. They also showed that the expression levels of IL-1β and IL-6 were higher in cells stimulated with CpG-ODN than those stimulated with Pam3CSK4 and LPS. These results suggested that TLR2, TLR4, and TLR21 in the monocytes of the cecal tonsil are functional and can recognize their specific ligands. However, it remains unknown whether the entire mucosal tissue containing monocytes as well as other cell populations, expresses cytokines in response to these TLR ligands.
Defensins are peptides that show antimicrobial activity against various microbes, including Gram-negative and Gram-positive bacteria, enveloped viruses, and fungi (Ganz et al., 2003). Zhang et al. (2016) detected gene expression of 11 out of 14 avian β-defensins (AvBDs) in the intestinal epithelial cells of chicken embryos, whereas Lu et al. (2015) observed the expression of seven AvBDs in the small intestinal mucosa of an 84-day-old chicken. Both studies revealed a synergistic effect of vitamin D3 with LPS-challenge in the induction of AvBD expression. These AvBDs may be co-expressed by different cells in the intestinal mucosa, as AvBD2 has been identified in mucosal leukocytes (Terada et al., 2018), and AvBD9 expression has been previously reported in enteroendocrine cells from the luminal and crypt epithelia (Cuperus et al., 2016). The expression of AvBD1, AvBD2, AvBD4, and AvBD6 in the cecum was increased by Salmonella typhimurium infection (Akbari et al., 2008), while AvBD2 expression was reduced by Salmonella enteritidis (SE) infection in cultured intestinal cells from a SE-susceptible chicken line (Derache et al., 2009). Su et al. (2017) reported that the response of AvBD expression to Eimeria challenge was inconsistent; E. acervulina downregulated six AvBDs in the chick small intestine, whereas E. maxima upregulated the expression of AvBDs in the small intestine and downregulated two AvBDs in the cecum. Thus, it is remains unknown whether the expression of AvBDs in the intestine is affected differentially after stimulation with different microbes in the intestine.
IL-1β and IL-6 are multifunctional proinflammatory cytokines responsible for modulating the functions of the immune system (Okada et al., 1983; Chomarat et al., 2000; Arend et al., 2008). Recently, we reported that IL-1β and IL-6 upregulated the AvBDs expressions in the chicken ovary and oviduct (Abdelsalam et al., 2012; Sonoda et al., 2013). Mathews et al. (1999) have also reported that IL-1β induces human β-defensin expression. Thus, a combined analysis of the expression levels of AvBDs and proinflammatory cytokines is important for understanding the defense system in the intestine against pathogenic agents.
To identify new strategies for improving the innate immune functions in the chick intestine, it is necessary to understand the effects of specific MAMPs on the expression of innate immune molecules. Thus, the aim of this study was to determine the effects of TLR2, TLR4, and TLR21 ligands on the expression of proinflammatory cytokines and AvBDs in the mucosal tissue of chick ileum and cecum. We utilized TLR2, TLR4, and TLR21 ligands to assess the response of innate immune factors to bacterial patterns, including substances derived from Gram-positive and Gram-negative bacteria, as well as unmethylated DNA. Among 14 AvBDs, AvBD1, 4, and 7 were selected for analysis as they showed clear polymerase chain reaction (PCR) products in our preliminary examination.
Broiler chicks (Chunky) were obtained by incubating fertilized eggs purchased from a local hatchery (Fukuda Breeders Co., Okayama, Japan). Male chicks were selected by feather discrimination and maintained in a brooding room with 24 h lighting for 3 days. They were provided a commercial starter diet (Nichiwa Sangyo Co. Ltd., Kobe, Japan) and water ad libitum. As the expression of AvBDs decreases from day 4 of life (Crhanava et al., 2011), three-day-old chicks were euthanized using carbon dioxide, and the ileum and cecum were collected. A part of the tissues were fixed in 10% (v/v) formalin in phosphate buffered saline (PBS), and processed for paraffin sectioning, followed by staining for histology. Other tissues were used for tissue culture to assess the effects of TLR ligands on cytokine and AvBD expression. This study was approved by the Hiroshima University Animal Research Committee (No. C15–16).
HistologyParaffin sections (4 µm) of the ileum and cecum were deparaffinized and stained using hematoxylin and eosin. They were dehydrated using ethanol, cleared in xylene, and covered. The sections were examined under a light microscope (Nikon Eclipse E400; Nikon Co., Tokyo, Japan).
Preparation of TLR LigandsPam3CSK4 (TLR2 ligand; a synthetic triacylated lipopeptide) was purchased from Novus Biologicals (Littleton, CO, USA). LPS from S. minnesota (TLR4 ligand) was purchased from Wako Pure Chemical Industries (Osaka, Japan). Class B CpG-ODN (2007) (TLR21 ligand), (5′-TCGTCGTTGTCGTTTTGTCGTT-3′) was purchased from InvivoGen (San Diego, CA, USA). These regents were dissolved in sterile water and stored at −20°C until use.
Incubation of Intestinal Tissue with TLR LigandsIntestinal tissue was cultured as described in previous studies (Abdel-Mageed et al., 2017; Kang et al., 2018), with minor modifications. The ileum and cecum from experimental birds were cut longitudinally and washed with PBS to remove the intestinal contents. They were then cut into small specimens (approximately 5 mm×5 mm) and each specimen was placed in a separate polystyrene tube (Bio-ONE Ltd., Tokyo, Japan) containing 4 ml incubation medium, which consisted of TCM-199 medium (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan) with 100 U/ml penicillin and 100 µg/ml streptomycin (Cosmo Bio Co., Ltd., Tokyo, Japan). They were incubated for 10 min at 39°C with 5% CO2 and 95% air in a CO2 incubator (Panasonic Co., Ltd., Osaka, Japan), followed by washing with incubation medium. Then, the intestinal specimens were placed in new polystyrene tubes containing 4 ml incubation medium. Different doses of TLR ligands were subsequently added into the medium to produce the following concentration ranges: Pam3CSK4 (0–100 ng/ml), LPS (0–1000 ng/ml), and CpG-ODN (0–100 ng/ml) (n=7 chicks in the Pam3CSK4 and LPS treatment groups; n=6 chicks in the CpG-ODN group). Then, the tissues were incubated for 1 or 3 h at 39°C with 5% CO2 and 95% air in a humidified CO2 incubator.
RNA Isolation and cDNA PreparationTotal RNA was extracted using Sepasol RNA I Super (Nacalai Tesque, Inc. Kyoto, Japan) per the manufacturer's instructions. The extracted total RNA samples were dissolved in TE butter (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) and stored at −80°C until further use. The RNA samples were treated with 1URQ1 RNase-free DNase (Promega Co. Madison, WI, USA) in a 10 µl reaction mixture (1 µg total RNA, 1× DNase buffer and 1UDNase) on a programmable thermal controller (PTC-100; MJ Research, Waltham, MA, USA), programmed at 37°C for 30 min and then at 65°C for 10 min with 1URQ1 DNase Stop Solution (Promega Co.). The concentration of RNA in each sample was measured using a NanoDrop Lite (Thermo Fisher Scientific, Waltham, USA). The RNA samples were then reverse-transcribed using ReverTra Ace (Toyobo Co. Ltd., Osaka, Japan) according to the manufacturer's instructions. The reaction mixture (10 µl) consisted of 0.5 µg total RNA, 1× reverse transcription buffer (Toyobo Co. Ltd.), 1 µM deoxyribonucleotide triphosphate (dNTP) mixture (Toyobo Co. Ltd.), 5U RNase inhibitor (Toyobo Co. Ltd.), 0.25 µg oligo(dT) 20 (Toyobo Co. Ltd.), and 50 UReverTra Ace. Reverse transcription was performed at 42°C for 30 min, followed by heat inactivation at 99°C for 5 min using a programmable thermal controller. The cDNA samples were stored at −20°C until use.
Real-time PCRReal-time PCR was performed using the AriaMix real-time PCR system (Agilent Technologies, Santa Clara, CA, USA). The reaction mixture (10 µl) consisted of 1µl cDNA, 1× Brilliant III SYBR Green QPCR mix (Agilent Technologies), 0.25 µM each forward and reverse primer, and 0.25 µM water. Table 1 shows the primer sequences used in this study. Two different thermal protocols were used for PCR, depending on the target genes, with either two or three steps. The two-step protocol was 50 cycles at 95°C for 5 s, followed by 60°C (RPS17), 62°C (AvBD4, 7, and IL-1β), or 63°C (IL-6) for 10 s. The three-step protocol for AvBD1 was 50 cycles at 95°C for 5 s, 55°C for 10 s, and 72°C for 10 s. Real-time PCR data was analyzed using the 2−ΔΔct method to calculate the relative level of gene expression in each sample and was expressed as ratios relative to the expression of the housekeeping gene RPS17 (Livak and Schmittgen, 2001). RNA samples from each control group (samples incubated without TLR ligand) were used as the standard.
| Gene | Sequences (5′-3′) | Accession No. (Reference) | |
|---|---|---|---|
| IL-1β | F | ACTGGGCATCAAGGGCTA | NM_204524 (a) |
| R | GGTAGAAGATGAAGCGGGTC | ||
| IL-6 | F | AGAAATCCCTCCTCGCCAAT | NM_204628 (b) |
| R | AAATAGCGAACGGCCCTCA | ||
| AvBD1 | F | GATCCTCCCAGGCTCTAGGAAG | NM_204993 (c) |
| R | GCCCCATATTCTTTTGC | ||
| AvBD4 | F | ATCGTGCTCCTCTTTGTGGCAGTTCA | NM_001001610 (c) |
| R | TACAACCATCTACAGCAAGAATACT | ||
| AvBD7 | F | ACCTGCTGCTGTCTGTCCTC | NM_001001194 (c) |
| R | TGCACAGCAAGAGCCTATTC | ||
| RPS17 | F | AAGCTGCAGGAGGAGGAGAGG | NM_204217 (b) |
| R | GGTTGGACAGGCTGCCGAAGT |
Reference for each primer is as follows; a=Zang et al. (2012), b=Nii et al. (2011), c=Xu et al. (2015), d=Subedi et al. (2007)
The values were presented as fold changes in the expression level of each gene compared to the control group in each chick (n=7 for Pam3CSK4 and LPS, n=6 for CpG-ODN). The statistical significance of the differences in the gene expression levels among different concentrations of TLR ligands was determined using the Kruskal-Wallis test, followed by the Steel test. Differences were considered significant when the P value was <0.05.
Figure 1 shows the histology of the ileum and cecum mucosa. The villi and crypts lined the luminal epithelium and crypt epithelia. Leukocytes, including small and round lymphocytes with densely stained nucleus and eosinophilic heterophil-like cells, were localized in the luminal and crypt epithelium and lamina propria. These structures were observed in both the ileum and cecum, although the villi were higher in the ileum than in the cecum (Fig. 1a and b, respectively).

Histology of the ileum (a) and cecum (b) of 3-d-old chicks. The villi (V) and crypt (Cr) are lined by luminal and crypt epithelium (E). Eosinophilic heterophil-like cells (arrow heads) are localized in the luminal and crypt epithelium and lamina propria (L) in both the ileum and cecum. Arrows show lymphocytes. Hematoxylin and eosin staining. Scale bars=50 µm.
Figures 2 and 3 show the effects of Pam3CSK4 on the expression of IL-1β, IL-6, and AvBDs in the ileum and cecum. In the ileum, IL-1β expression tended to be downregulated by 3 h incubation with 1 ng/ml Pam3CSK4 (P<0.05; Fig. 2b). The expression levels of AvBD1 and AvBD7 were also reduced by incubation with 1 to 100 ng/ml Pam3 CSK4 for 1 h (P<0.05; Fig. 2e, i). The expression levels of IL-6 and AvBD4 were not affected by Pam3CSK4 in the ileum (Fig. 2). In contrast, in the cecum, IL-1β expression tended to increase after incubation with 100 ng/ml Pam3CSK4 for 1 h (Fig. 3a). AvBD1 expression was upregulated by 3 h incubation with 100 ng/ml Pam3CSK4, and AvBD7 expression was also increased by incubation with 1 to 100 ng/ml Pam3CSK4 for 3 h (P<0.05; Fig. 3f, j). Incubation with Pam3CSK4 did not significantly affect the expression levels of IL-6 and AvBD4 in the cecum (Fig. 3).

Changes in the expression of proinflammatory cytokines and AvBDs in the cultured chick ileum in response to Pam3CSK4 stimulation. Ileum tissues were cultured with 1 (●), 10 (▲), or 100 (■) ng/ml Pam3CSK4 for 1 or 3 h (n=7 for each condition). The values of the dots show the fold changes in gene expression (Y axis is presented by a logarithmic scale) in Pam3CSK4-treated tissue relative to non-treated tissue (0 ng/ml Pam3CSK4; value=1, dotted line). The solid bar represents the median value within each treatment. Asterisks indicate significant differences between treated and non-treated groups, determined via Kruskal-Wallis and Steel tests (* P<0.05, ** P<0.01).

Changes in the expression of proinflammatory cytokines and AvBDs in the cultured chick cecum in response to Pam3CSK4 stimulation. Cecum tissues were cultured with 1 (●), 10 (▲), or 100 (■) ng/ml Pam3CSK4 for 1 or 3 h (n=7 for each condition). The values of the dots show fold changes in gene expression in Pam3CSK4-treated tissue relative to non-treated tissue (0 ng/ml Pam3CSK4; value=1, dotted line)). See Fig. 2 for the other experiment. *=Significantly different at P<0.05 between treated and non-treated groups.
The effects of LPS stimulation on the expression levels of IL-1β, IL-6, and AvBDs in the ileum and cecum are shown in Figs. 4 and 5, respectively. In the ileum, IL-1β expression was increased by incubation with 1000 ng/ml LPS for 1 h (p<0.05; Fig. 4a), but was lowered by incubation with 10 or 100 ng/ml LPS for 3 h (P<0.05; Fig. 4b). Incubation with 10 ng LPS for 3 h reduced the expression levels of IL-6 and AvBD1 (P<0.05; Fig. 4d, f). LPS did not significantly affect the expression levels of AvBD4 and AvBD7 in the ileum (Fig. 4). In the cecum, the expression levels of IL-1β and IL-6 tended to decrease in response to 3-h incubation with 10 or 1000 ng/ml LPS (P<0.05; Fig. 5b, d). However, the expression levels of AvBD1 and AvBD7 tended to be upregulated by incubation with 100 or 1000 ng/ml LPS for 1 h, respectively (P<0.05; Fig. 5e, i), and the expression level of AvBD4 was increased by incubation with 100 ng/ml LPS for 3 h (P<0.05; Fig. 5h).

Changes in the expression of proinflammatory cytokines and AvBDs in the cultured chick ileum in response to LPS stimulation. Ileum tissues were cultured with 10 (●), 100 (▲), or 1000 (■) ng/ml LPS for 1 or 3h (n=7 for each condition, except for AvBD4 data of 3 h incubation with 100 ng/ml LPS (n=6), in which one bird data was omitted as its expression was not detectable.). The values of the dots show the fold changes in gene expression in LPS-treated tissue relative to non-treated tissue (0 ng/ml LPS; value=1, dotted line). See Fig. 2 for the other experiment. *=Significantly different at P<0.05 between treated and non-treated groups.

Changes in the expression of proinflammatory cytokines and AvBDs in the cultured chick cecum in response to LPS stimulation. Cecum tissues were cultured with 10 (●), 100 (▲), or 1000 (■) ng/ml LPS for 1 or 3h (n=7 for each condition). The values of the dots show the fold changes in gene expression in LPS-treated tissue relative to non-treated tissue (0 ng/ml LPS; value=1, dotted line). See Fig. 2 for the other experiment. *=Significantly different at P<0.05 between treated and non-treated groups.
Figures 6 and 7 show the effects of CpG-ODN on the expression levels of IL-1β, IL-6, and AvBDs in the ileum and cecum, respectively. In the ileum, the expression level of IL-1β was decreased by incubation with 0.1 or 10 ng/ml CpGODN for 3 h (P<0.01; Fig. 6b), whereas the expression level of IL-6 was markedly upregulated at approximately 5,000-fold by incubation with 10 ng/ml CpG-ODN for 3 h (p<0.01; Fig. 6d). The expression levels of AvBD1 and AvBD4 were lowered by incubation with CpG-ODN (P<0.01, Fig. 6e, f, g). However, incubation with 1 ng/ml CpG-ODN for 3 h markedly upregulated the expression level of AvBD7 by approximately 400-fold (P<0.01; Fig. 6j). In the cecum, the expression level of IL-1β was increased by incubation with 0.1 or 1 ng/ml CpG-ODN for 3 h (P<0.01; Fig. 7b), whereas CpG-ODN did not significantly affect the expression of other target genes, namely, IL-6, AvBD1, AvBD4, and AvBD7 (Fig. 7).

Changes in the expression of proinflammatory cytokines and AvBDs in the cultured chick ileum in response to CpG-ODN stimulation. Ileum tissues were cultured with 0.1 (●), 1 (▲), or 10 (■) ng/ml CpG-ODN for 1 or 3 h (n=6 for each condition). The values of the dots show the fold changes in gene expression in CpG-ODN-treated tissue relative to non-treated tissue (0 ng/ml CpG-ODN; value=1, dotted line). See Fig. 2 for other explanations. **=Significantly different at P < 0.01 between treated and non-treated groups.
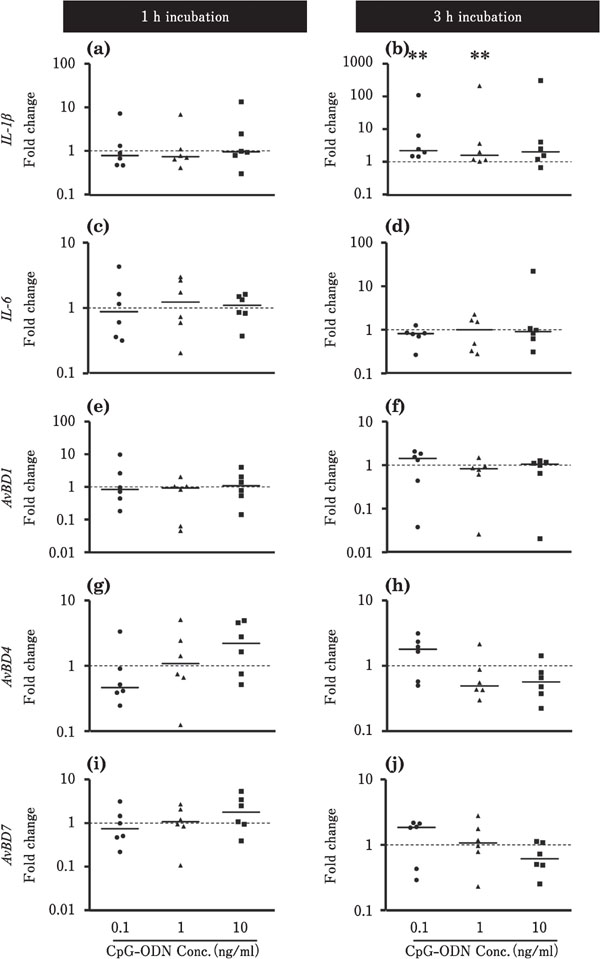
Changes in the expression of the proinflammatory cytokines and AvBDs in the cultured chick cecum in response to CpG-ODN stimulation. Cecum tissues were cultured with 0.1 (●), 1 (▲), or 10 (■) ng/ml CpG-ODN for 1 or 3h (n=6 for each condition). The values of the dots show the fold changes in gene expression in CpG-ODN-treated tissue relative to non-treated tissue (0 ng/ml CpGODN; value=1, dotted line). See Fig. 2 for other explanations. **=Significantly different at P<0.01 between treated and non-treated groups.
We report herein that TLR2, TLR4, and TLR21 ligands, specifically Pam3CSK4, LPS, and CpG-ODN, modulate the expression of proinflammatory cytokines (IL-1β and IL-6) and three AvBDs in cultured chick intestinal tissues. Our major findings were as follows: (1) Pam3CSK4 downregulated IL-1β, AvBD1, and AvBD7 in the ileum, but upregulated them in the cecum; (2) LPS downregulated IL-1β and IL-6 expression in both the ileum and the cecum, whereas it upregulated all AvBDs in the cecum; (3) CpG-ODN downregulated the expression levels of IL-1β, AvBD1, and AvBD7 in the ileum, and significantly upregulated the expression levels of IL-6 and AvBD7 in the ileum and that of IL-1β in the cecum.
Our results suggested that the mucosal tissues of the ileum and cecum, which contain leukocytes, including heterophil-like cells, luminal and crypt epithelial cells, and other enterocytes, express not only IL-1β and IL-6, but also AvBD1, 4, and 7. These observations support the results of previous reports, showing that the AvBD genes were expressed in the small intestine and cecum of embryos and chicks (Akbari et al., 2008; Crhanova et al., 2011; Hong et al., 2012; Lu et al., 2015; Zang et al., 2016; Su et al., 2017; Terada et al., 2018). AvBD2 in the mucosal leukocytes and AvBD9 in enteroendocrine cells from the luminal and crypt epithelia have also been reported (Cuperus et al., 2016; Terada et al., 2018). Generally, we did not observe clear dose dependencies in the response of many cytokines and AvBDs to TLR ligands in this study. Although the precise reason for this observation is not known, we assume that it was due to the relatively small changes in their expression in response to TLR ligands.
Makinnon et al. (2009) reported that TLR1-1, TLR2-1, TLR2-2, TLR3, TLR4, TLR5, TLR7, TLR15, and TLR21 were expressed in the small and large intestines of chicks, and with the exception of TLR21, the distal segments expressed more TLR than the proximal segments. Thus, it is expected that different MAMPs are recognized by these TLRs in the intestinal mucosa cells. In this study, stimulation of the cecum with Pam3CSK4 and the ileum with LPS for 1 h upregulated IL-1β (Fig. 3a, 4a). CpG-ODN stimulation upregulated IL-6 in the ileum (Fig. 6d) and IL-1β in the cecum (Fig. 7b). Our results suggest that infection by Gram-positive and Gram-negative bacteria induce the expression of IL-1β and IL-6 in the chick intestine via the recognition of their pattern molecules by TLRs. The synthesized IL-1β and IL-6 probably regulate immune response as they are multifunctional cytokines that modulate the immune defense system (Okada et al., 1983; Chomarat et al., 2000; Arend et al., 2008). In particular, IL-6 expression markedly increased in the ileum in response to CpG-ODN (Fig. 6d), suggesting that IL-6 expression is upregulated by DNA-bearing microbes such as bacteria and DNA viruses. Although many leukocytes may not be mature in 3-day-old chicks, IL-6 synthesized in the ileum in response to CpG-ODN of luminal microbes may stimulate their differentiation and maturation in the mucosa.
Taha-abdelaziz et al. (2016) also reported that stimulation with Pam3CSK4, LPS, and CpG-ODN upregulated IFNα, IL-1β, IL-6, and CXCLi2 in cecum tonsil monocytes. However, our results revealed an inhibitory effect of Pam3CSK4 on IL-1β expression in the ileum, as well as an inhibitory effect of LPS on IL-1β and IL-6 expression in the ileum and cecum after 3 h incubation. Although the reason for these inhibitory effects were identified in the current study unlike the report of Taha-abdelaziz et al. (2016) is unknown, we assume that the expression of these cytokines in the monocytes isolated from cecum tonsil differed from that of the whole mucosal tissue used in our study. Although we did not use isolated specific cells in this study, the cytokine response of the whole mucosal tissue to MAMPs may reflect the actual cytokine expression profile in mucosa colonized or infected by microbes.
In the cecum, we observed upregulation of AvBD1 and AvBD7 by Pam3CSK4 (Fig. 3f, j) and upregulation of AvBD1, AvBD4, and AvBD7 by LPS (Fig. 5e, h, i). Taha-abdelaziz et al. (2016) reported that in isolated cecal tonsil mononuclear cells whose AvBD1, AvBD2, and cathelicidin 3 expression was not affected by Pam3CSK4, LPS, and CpG-ODN (Taha-abdelaziz et al., 2016). We suggest that, unlike the isolated mononuclear cells, in the cecum mucosa – containing the entire mucosal cell population – AvBD expression may be induced by the interaction of TLR2 and TLR4 with Pam3CSK4 and LPS of Gram-positive and Gram-negative bacteria.
In contrast to that in the cecum, the expression levels of AvBD1 and AvBD7 in the ileum were downregulated or were not affected by stimulation with Pam3CSK4 (Fig. 2e, i). The abundance or diversity of intestinal bacteria increases during the first weeks of life in chicks (Crhanova et al., 2011, Danzeisen et al., 2011). Among these bacteria, the small intestine is dominated by Lactobacillus species, namely Gram-positive bacteria (Gong et al., 2007; Konsak et al., 2013; Kers et al., 2018). Akbari et al. (2008) reported that although the Salmonella typhimurium challenge upregulated AvBD expression, feeding chicks with probiotics such as Lactobacillus acidophilus suppressed the increase in AvBD expression. The downregulation of AvBDs by Pam3CSK4 may explain which AvBD synthesis in response to luminal microbes is suppressed by Gram-positive bacteria, probably due to recognition of their pattern molecule by TLR2 in the ileum.
In the ileum, CpG-ODN stimulation downregulated AvBD1 and AvBD4 (Fig. 6e, f, g), but significantly upregulated AvBD7 expression by approximately 400-fold (Fig. 6j). These results suggest that bacterial and viral DNA affect the expression of AvBDs via TLR21 in the ileum mucosa. In particular, their effect on the expression of AvBD7, which is markedly increased by CpG-ODN stimulation, may be important for preventing infection with pathogenic agents containing unmethylated DNA. We have previously reported that IL-1β and IL-6 induced AvBD expression in the chicken ovary and oviduct (Abdelsalam et al., 2012; Sonoda et al., 2013). The present study showed that CpG-ODN induced markedly high expression of IL-6 (5000-fold change) and AvBD7 (400-fold change) in the ileum (Fig. 6d, j). Thus, IL-6 may support AvBD7 synthesis in the ileum, although further studies are necessary to confirm this. On the other hand, CpG-ODN did not significantly affect AvBD expression in the cecum. Although the reason for this remains unknown, the cecum mucosa may be less sensitive to CpGODN due to exposure to specific bacterial substances in the cecum lumen where the microbe complex is rich.
In conclusion, we suggest that the expression of proinflammatory cytokines (IL-1β and IL-6) and AvBDs (AvBD1, AvBD4, and AvBD7) in the ileum and cecum is affected by TLR2, TLR4, and TLR21 ligands. Although the specific cell types expressing these cytokines and AvBDs remain to be identified, the response of the whole pieces of the intestinal mucosa to the MAMPs may reveal the realistic local immunodefense function in the mucosa in situ. The types of cells expressing AvBDs may differ between the ileum and cecum, as AvBD expression was downregulated by Pam3CSK4 and LPS, whereas it was upregulated in the cecum. Studies on the direct effects of MAMPs on the expression of innate immune molecules in the chick intestine were previously limited. The novel findings of this study will be important for understanding the relationship between immune factor expression and the intestinal microbe complex.
This work was supported by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (No. 17H03904) to YY.
The authors declare that there are no conflicts of interest.