2025 Volume 62 Article ID: 2025009
2025 Volume 62 Article ID: 2025009
Abstract: Salmonella enterica and coccidia (Eimeria spp.) are important intestinal pathogens in broiler production. Salmonella has high zoonotic potential, and coccidia are responsible for large economic losses. Live vaccines reduce shedding of Salmonella and minimize the impact of coccidial infections on broiler performance. This study investigated the interaction between both vaccines on the intestinal health of broilers. The 2 × 2 experimental design included vaccination against Salmonella Typhimurium (ST) (no vaccination or vaccination on day 14) and vaccination against coccidiosis (no vaccination or vaccination on day 1). On day 28, all groups were challenged with a ST marker strain resistant to nalidixic acid. Re-isolation of ST from the liver and ceca on day 42 indicated higher susceptibility to systemic infection with ST in birds vaccinated against coccidiosis than that in unvaccinated birds. On day 42, cecal immunoglobulin A (IgA) levels against ST decreased in the group vaccinated against ST and coccidia compared to those in all other groups. IgG antibodies in the cecal contents significantly decreased in the group vaccinated against coccidiosis compared to that of the group vaccinated against ST. There was no difference in systemic IgG levels among groups. Analysis of the cecal microbiota revealed a significant difference in beta diversity on days 28 and 42 between the groups vaccinated against coccidiosis and unvaccinated groups. Functional pathway profiling showed increased activity of pathways associated with carbohydrate and arachidonic acid metabolism in the group vaccinated against ST compared to that in other groups. Gene expression of claudin 1, claudin 4, E-cadherin, β-catenin, and zonula occludens 2 in the cecal wall differed between the groups on days 28 and 42. These findings indicated the significant influence of ST and coccidiosis vaccines on the intestinal health of broilers; however, further studies are required to clarify the implications for health and performance.
Salmonella is one of the most common pathogens that cause food-borne diseases in humans[1]. In the U.S., more than 1.2 million human illnesses and 450 deaths are related to salmonellosis each year; in Europe, 91,857 confirmed cases were reported in 2018[2,3]. Broiler meat is an important vector of Salmonella infection, and Salmonella Typhimurium (ST) is one of the most commonly isolated serovars in the U.S. and Europe[2,4]. An important strategy to control disease in humans and minimize the risk of meat contamination is vaccination of chickens. Attenuated live and inactivated vaccines against ST are commercially available[5].
Live vaccines are widely used in the broiler chicken industry. These vaccine strains colonize the intestinal mucosa and stimulate a mucosal immune response[6,7]. This is especially relevant for pathogens, such as Salmonella and coccidia, which tend to remain in the intestinal tract[8,9]. In addition, live vaccines are suitable for mass application, which is important for treating large broiler flocks. In contrast, inactivated vaccines are administered to broiler breeders and layers, often in combination with live vaccines, to induce a strong and long-lasting antibody response that reduces egg and cecal colonization, shedding, and transmission[6,10,11].
Coccidiosis is a gastrointestinal disease in young chickens that causes economic losses in poultry production of up to $1,528 million in the U.S. alone[12]. Coccidia cause this damage by invading the intestinal mucosa, replicating in the enterocytes, and thereby inducing epithelial cell damage and inflammation[13,14]. Live vaccines against coccidiosis, containing sporulated oocysts of Eimeria tenella, Eimeria maxima, Eimeria acervulina, and other Eimeria spp., are frequently administered in the hatchery[15,16].
Previous studies have demonstrated multiple interactions between Salmonella spp. and Eimeria spp. in chickens. Co-infection with E. tenella or E. necatrix and ST led to increased ST growth in the ceca of birds housed in cages[17,18,19,20]. When young chickens were infected with E. tenella, cage contamination with ST resulted in a higher probability of acute salmonellosis than that in controls that were not infected with E. tenella[21]. Chickens in floor pens infected with E. tenella, E. acervulina, or E. maxima took longer to clear an ST infection than chickens receiving anticoccidial medication. Additionally, coccidiosis predisposes chickens to systemic salmonellosis[22,23]. In contrast, a field study found that administration of live sporulated Eimeria oocysts in hatcheries reduced Salmonella spp. prevalence throughout the production cycle[24].
Since coccidiosis vaccines and live attenuated vaccines against ST may be administered simultaneously in hatcheries, an interaction between the two vaccines is likely; however, to our knowledge, this has never been investigated[11,25]. Therefore, this pilot study tested the effect of a coccidiosis vaccine on the efficacy of ST vaccination and the effect of both vaccines on the intestinal health of broilers, as defined by the complex interaction of local immunity, intestinal microbiota, and intestinal wall integrity.
The animal study protocol was approved by the Auburn University Institutional Animal Care and Use Committee (protocol 2019-3638).
One hundred and sixty unvaccinated and unsexed day-old broiler chicks were obtained from a commercial hatchery and randomly placed in four floor pens. Each pen measured 2.8 m2 and contained 40 birds. The experimental setup consisted of four groups, each housed in one pen, with different treatments as shown in Table 1. On the first day, two groups (C and CST) were administered one dose of a coccidiosis vaccine containing E. acervulina, E. maxima, and E. tenella (Advent, Huvepharma, Peachtree City, GA, USA) via oral gavage. The other two groups were not vaccinated against coccidiosis (Control [Ctrl] and ST) and were administered 0.02% coccidiostat nicarbazin (Phibro, Teaneck, NJ, USA) in the feed during the entire study to prevent infection with coccidia. At 14 days of age, the CST and ST groups received one dose of a commercial ST vaccine (Poulvac ST, Zoetis, Parsippany, NJ, USA) via crop gavage. All birds were fed a standard broiler diet formulated to meet or exceed the National Research Council recommended minimum nutrient requirements for broilers[26] throughout the experiment and had unlimited access to feed and water.
| Group1 | Coccidiosis vaccine (day 1)2 | ST vaccine(day 14) |
| Ctrl | No | No |
| C | Yes | No |
| ST | No | Yes |
| CST | Yes | Yes |
1 Group names: Control (Ctrl), unvaccinated; C, vaccinated only with coccidia on day 1; ST, vaccinated only with Salmonella Typhimurium (ST) on day 14; CST, vaccinated with both coccidia on day 1 and ST on day 14. All groups were challenged with the ST marker strain on day 28.
2 Groups not vaccinated against coccidiosis (Ctrl and ST) were fed a diet containing 0.02% nicarbazin throughout the entire study.
On day 28, 20 birds per group were randomly selected and euthanized, with 20 birds remaining in each pen. Euthanasia was performed using carbon dioxide followed by cervical dislocation. The cecal wall, cecal content, and blood were collected from 15 birds for the detection of antibodies, characterization of cecal microbiota, and expression of selected genes in the intestinal wall. On the same day, all remaining birds were challenged with 1 mL brain heart infusion (BHI) broth containing 104 colony-forming units (cfu) of the ST marker strain by oral gavage. On day 42, the birds were euthanized for sampling, as described above. In addition, cecal tonsils and livers were collected for the re-isolation of ST.
Preparation of ST inoculumThe ST marker strain used in this study was resistant to nalidixic acid. The stock was stored in glycerol at −80°C. A sample of this stock was streaked out on trypticase soy II agar with 5% sheep blood (BD Difco, Franklin Lakes, NJ, USA) and incubated for 24 h at 37°C. BHI broth (BD Difco) was inoculated with one typical colony and incubated overnight, shaking at 200 rpm at 37°C. Based on the counts acquired from an overnight culture of one ST colony from a previous study, the inoculum was diluted to approximately 104 (cfu/mL) to use in the challenge[27]. To confirm the strength of the inoculum, a ten-fold dilution series was plated on xylose lysine tergitol-4 (XLT4) agar plates (Criterion, Hardy Diagnostics, Santa Maria, CA, USA) containing 100 µg/mL nalidixic acid. Colonies were counted after incubation for 24 h at 37°C.
Re-isolation of STOn day 42 of the experiment, cecal tonsils and 1 g of the liver were sampled from 15 chickens per group. The organs were incubated in tetrathionate broth with 20 mL iodine solution/L (BD Difco) overnight at 37°C. The next day, samples were streaked on XLT4 agar plates containing 100 µg/mL nalidixic acid. After an incubation period of 24 h at 37°C, the plates were checked for the presence of black colonies, indicative of Salmonella, using a qualitative approach (presence/absence only).
Detection of ST antibodies in blood and cecal contentOn both sampling days (day 28 and day 42), 1–1.5 mL blood was collected from 10 birds per group by puncture of the brachial wing vein. The tubes were maintained at room temperature for 1 h and centrifuged at 3000 rpm (2143 × g) for 10 min. The obtained serum was stored at −20°C.
Cecal content from 10 birds per group was diluted 1:2 with proteinase inhibitor solution containing 0.15 M NaCl, 0.005 M NaH2PO42H2O, 0.005 M Na2HPO4, 0.02% sodium azide, 0.005 M EDTA-Na2H2O, and 0.002 M phenylmethylsulfonyl fluoride (Q-1395, Bachem AG, Bubendorf, Germany), and immediately stored on ice. The tubes were centrifuged at 3000 rpm (2143 × g) for 10 min at 5°C. The supernatant was harvested and stored at −20°C.
Serum and cecal anti-ST immunoglobulin G (IgG) levels were detected using an indirect ELISA kit for Salmonella Enteritidis-Typhimurium antibodies (BioChek, Kings Ride, Ascot, Berkshire, UK), following the manufacturer’s instructions.
To optimize detection of IgA in cecal content, two checkerboard analyses were performed to determine an optimal protocol. First, 1:10, 1:50, and 1:100 dilutions of cecal content, and 1:1000, 1:2000, and 1:5000 dilutions of alkaline phosphatase- labeled rabbit anti-chicken IgA antibodies (Bio-Rad, Hercules, CA, USA) as secondary antibodies were tested with one sample of the control group (day 28) as a negative control and one sample of the control group after the ST challenge (day 42) as a positive control. The second checkerboard analysis was performed with the same sample dilutions, but used combinations of two antibodies at different concentrations: a goat anti-chicken IgA antibody at 1:1000 and 1:2000 dilutions and an alkaline phosphatase- labeled rabbit anti-goat IgG antibody (Bethyl Laboratories, Montgomery, TX, USA) at 1:5000 and 1:10 000 dilutions, to enhance the optical signal.
The largest difference between negative and positive samples was observed with a sample dilution of 1:50 and an alkaline phosphatase-labeled goat anti-chicken IgA dilution of 1:1000 (results not shown). Therefore, these dilutions were selected to test all cecal samples for IgA antibodies.
Cecal microbiome analysisOn both sampling days (days 28 and 42), cecal content was collected from three birds in each group. Following the manufacturer’s instructions, DNA was extracted from 0.2 g cecal content using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). Between 400 and 450 base pairs of bacterial 16S rDNA were amplified by polymerase chain reaction (PCR) using a Taq PCR Master Mix Kit (Qiagen). For the amplification, 2 µL template DNA, 10 pmol forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) with the linker sequence CS1 and 10 pmol reverse primer 926R (5′-CCGYCYYTTYMTTTRAGTTT-3′) with the linker sequence CS2 were used in a total volume of 25 µL[28,29]. The PCR conditions were as follows: an initial denaturation step of 3 min at 94°C, followed by 28–40 cycles of denaturation for 30 s at 94°C, annealing at 50°C for 30 s, extension at 72°C for 60 s, and a final elongation at 72°C for 10 min. PCR products were observed using agarose gel electrophoresis, purified, and sent to the University of Illinois, Chicago DNA Service Facility for targeted amplicon sequencing on an Illumina MiSeq platform[30]. The resulting demultiplexed paired-end reads were imported into qiime2 2023.2[31]. The Dada2 plugin was used to trim 19 nucleotides of the forward reads and 20 nucleotides of the reverse reads, based on the given primer sequence lengths, and to truncate the sequences to a length of 250 nucleotides[32]. The sequences were denoised and amplicon sequence variants were obtained. The metadata sheet was written on Google sheets, validated by Keemei[33], and imported into qiime2. The taxonomic composition of the bacterial communities was assessed using a Naïve Bayes classifier trained on 99% full-length sequences sourced from the SILVA (version 138) ribosomal RNA gene database project[34].
The data were imported into R using the qiime2R package version 0.99.6, and subsequent analysis was conducted using the phyloseq package version 3.17 and vegan package version 2.5-7[35,36,37]. All non-bacterial taxa were excluded from further analysis and samples containing less than 5000 reads were removed from the dataset to ensure data quality. Additionally, features with abundance below 0.01 were filtered out to enhance the visualization of the relative abundance.
Pathway analysis was performed using picrust2 version 2.5.1[38]. The output was imported into R, and differential analysis and visualization were conducted using ggpicrust2 1.7.0 and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database[39]. Raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under accession number PRJNA1011958.
Relative gene expression in the cecal wallOn days 28 and 42, a piece of the cecal wall, approximately 1 cm upstream of the cecal apex, was collected from five broilers per group and stored in 1 mL RNA stabilization solution (Qiagen). After 24 h at 4°C, the supernatant was discarded and the samples were frozen at −80°C. RNA was extracted from 15 mg of tissue using an RNeasy kit, according to the manufacturer’s instructions (Qiagen). The procedure included DNA digestion using Qiagen RNase-Free DNase (Qiagen). Based on the concentration of RNA determined by spectrophotometry using a Nanodrop 1000 (Thermo Scientific, Waltham, MA, USA), the samples were diluted with nuclease-free water to approximately 1 µg/µL RNA. Total RNA was transcribed into cDNA using a LunaScript RT SuperMix Kit (New England Biolabs, Ipswich, MA, USA). The thermal program consisted of 2 min at 25°C for primer annealing, 10 min at 55°C for cDNA synthesis, and 1 min at 95°C for heat inactivation.
Two technical replicates containing 2 µL cDNA of each sample were amplified by quantitative PCR (qPCR) using the PerfeCta SYBR Green FastMix (QuantaBio, Beverly, MA, USA) and primers for the genes of interest (claudin 1, 2, 4, 5; zonula occludens 1, 2; E-cadherin; and β-catenin) and the reference genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hydroxymethylabilane synthase (HMBS) (Sigma-Aldrich, Darmstadt, Germany). Detailed primer sequences are provided in Table 2. The cycling parameters for each gene were standardized and included the following steps: an initial denaturation cycle at 95°C for 10 min, followed by 40 cycles of denaturation (30 s at 95°C), annealing (1 min at 60°C), and extension (30 s at 72°C). PCR was performed using qTower3G (Analytik Jena, Jena, Germany). To calculate primer efficiency, a standard curve for each primer was performed using sample dilutions of 1, 1:10, 1:100 and 1:1000, with two technical replicates each. The slope was calculated using the slope function in Microsoft Excel based on the average Ct value and log quantity range. The primer efficiency was calculated using the formula:
| Target gene | Primer sequences (5′-3′) | PCR efficiency | Amplicon length (base pairs) | Reference |
| Claudin 1 (CLDN1) | F: CTG ATT GCT TCC AAC CAG R: CAG GTC AAA CAG AGG TAC AAG1 |
1.86 | 662 | [40] |
| Claudin 2 (CLDN2) | F: CCT CAG CCC TCC ATC AAA R: CTG CGT CTT CTC CTC TTA CTGT |
2.04 | 162 | [41] |
| Claudin 4 (CLDN4) | F: GAA GCG CTG AAC CGA TAC CA
R: TGC TTC TGT GCC TCA GTT TCC |
1.85 | 137 | [42] |
| Claudin 5 (CLDN5) | F: CAT CAC TTC TCC TTC GTC AGC
R: GCA CAA AGA TCT CCC AGG TC |
1.95 | 224 | [40] |
| Zonula occludens 1 (ZO1) | F: CTT CAG GTG TTT CTC TTC CTC CTC
R: CTG TGG TTT CAT GGC TGG ATC |
1.98 | 101 | [43] |
| Zonula occludens 2 (ZO2) | F: CGG CAG CTA TCA GAC CAC TC
R: CAC AGA CCA GCA AGC CTA CAG |
2.02 | 89 | [44] |
| E-cadherin (Ecad) | F: TCA CGG GCA GAT TTC TAT R: CAC GGA GTT CGG AGT TTA |
1.92 | 109 | [45] |
| β-catenin (Bcat) | F: CTG TTC AGA ATG TCG GAG GA
R: CTG GGC ACC AAT GTC AAG |
1.95 | [46] | |
| GAPDH | F: TGG AGA AAC CAG CCA AGT AT
R: GCA TCA AAG GTG GAG GAAT |
2.07 | 145 | [46] |
| HMBS | F: GAT GGA TCC GAT AGC CTG AA
R: GAT GTG CTT AGC TCC CTT GC |
2.01 | 195 | [47] |
1 F = forward, R = reverse. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HMBS, hydroxymethylabilane synthase.
Relative gene expression (RGE) was calculated using the Vandesompele method with two reference genes (REF) and eight genes of interest (GOI) and considering primer efficiencies (E) using the formula
All statistical tests were performed using R version 4.3.1[50] with a critical value of α = 0.05. Non-parametric methods were used when the dataset did not meet the assumptions for parametric testing.
Re-isolation rates were compared between groups using the chi-square test and logistic binomial regression, followed by pairwise comparison with Tukey’s correction.
Statistical analysis of differences in the sample to positive (S/P) ratio of immunoglobulins between groups was performed using the Kruskal–Wallis test and Dunn’s post hoc test with Bonferroni adjustment.
For the evaluation of bacterial composition, the taxonomic levels tested were phylum, family, and genus. The test groups were sorted by treatment, coccidia vaccination, or ST vaccination. Alpha diversity was analyzed using Kruskal–Wallis and Dunn’s post hoc tests with Bonferroni adjustment based on Shannon diversity, Simpson evenness, and observed richness. Beta diversity was analyzed using permutational multivariate analysis of variance (PERMANOVA) and pairwise PERMANOVA with Bonferroni adjustment, based on Jaccard distance, Bray–Curtis distance, and weighted and unweighted UniFrac distances. Analysis of compositions of microbiomes with bias correction was performed using qiime2 to test for differential abundance[51].
Differential abundance analysis of functional pathways was performed using the ANOVA-like differential expression (ALDEx) package ALDEx2 with the Kruskal–Wallis test, Benjamini Hochberg adjustment, and a generalized linear model[39,52,53].
For statistical analysis of relative gene expression, the data were base 2 log transformed, followed by Kruskal–Wallis and pairwise Wilcoxon rank sum tests with Bonferroni adjustment for outliers and a skewed distribution.
The ST challenge strain was re-isolated from five cecal tonsil samples from birds in the unvaccinated control group but not from liver samples. In birds vaccinated only against coccidia (group C), ST was re-isolated from two cecal tonsils and three liver samples. In the group that was vaccinated only against ST (group ST), one cecal tonsil sample was positive for ST, and no ST was detected in the liver samples. In birds vaccinated against both pathogens (group CST), two cecal tonsil samples and one liver sample were positive for ST. In summary, the lowest number of ST-positive samples was observed in birds vaccinated only against ST, and re-isolation of the ST challenge strain from the liver was successful only in birds vaccinated against coccidia, with or without ST vaccination. The control group showed active ST infection in the ceca, with no systemic spread to other organs (Table 3). Counts between groups were not significantly different.
| Group1 | Cecal tonsils | Liver |
| Ctrl | 5/152 | 0/15 |
| C | 2/15 | 3/15 |
| ST | 1/15 | 0/15 |
| CST | 2/15 | 1/15 |
1 Group names: Control (Ctrl), unvaccinated; C, vaccinated only with coccidia on day 1; ST, vaccinated only with Salmonella Typhimurium (ST) on day 14; CST, vaccinated with both coccidia on day 1 and ST on day 14. All groups were challenged with the ST marker strain on day 28.
2 Positive samples/Tested samples.
Anti-ST IgG antibodies were observed in both blood serum and cecal contents, whereas IgA antibodies were observed only in the cecal content. Statistical analysis revealed no significant differences in the IgG and IgA antibody responses between the experimental groups on day 28. On day 42, 14 days after challenge with ST, there were no differences in serum IgG antibodies between the groups. IgA antibodies in the cecal contents were significantly lower in birds vaccinated against coccidiosis and ST (group CST) than those in all other groups (Table 4). Levels of IgG antibodies in the cecal contents of birds vaccinated only against coccidiosis (group C) were significantly lower than those in the group that received only the ST vaccine (group ST).
| Group1 | Day 28 | Day 42 | ||||
| IgA in cecal content | IgG in cecal content | IgG in serum | IgA in cecal content | IgG in cecal content | IgG in serum | |
| Ctrl1 | 0.643 (0.391)2 |
0.024 (0.025) |
0.017 (0.008) |
0.500A
(0.270) |
0.021AB
(0.010) |
0.113 (0.073) |
| ST | −0.0714 (0.416) |
−0.001 (0.002) |
0.013 (0.008) |
0.320A
(0.085) |
0.061A
(0.025) |
0.062 (0.031) |
| C | −0.400 (0.213) |
0.012 +0.006) |
0.012 (0.004) |
0.360A
(0.181) |
0.027B
(0.024) |
0.118 (0.042) |
| CST | −0.171 (0.308) |
0.060 (0.033) |
0.009 (0.005) |
−0.580B
(0.144) |
0.026AB
(0.011) |
−0.007 (0.022 |
1 Group names: Control (Ctrl), unvaccinated; C, vaccinated only with coccidia on day 1; ST, vaccinated only with Salmonella Typhimurium (ST) on day 14; CST, vaccinated with both coccidia on day 1 and ST on days 14. All groups were challenged with the ST marker strain on day 28.
2 Mean and standard error of sample to positive (S/P) ratio.
Different superscripts indicate significant differences between groups (P ≤ 0.05).
Before filtering, 677 taxa were detected in the 12 samples collected on day 28, and 952 taxa in the 12 samples collected on day 42. Filtering based on the bacterial kingdom resulted in the detection of 638 taxa on day 28 and 877 taxa on day 42. Four samples from day 28 and one sample from day 42 had less than 5000 read counts and were excluded from the analysis (one Ctrl, two C, and one ST on day 28, and one Ctrl on day 42).
Analysis of the relative abundance on day 28 revealed that the phylum Firmicutes exhibited the highest relative abundance across all groups, followed by Bacteroidota, Cyanobacteria, Proteobacteria, and Desulfobacterota (Fig.1A). Among families, the most abundant were Clostridia_vadin_BB60 group, Bacteroidaceae, Rikenellaceae, Ruminococcaceae, and Oscillospiraceae (Fig. 1B). The genera with the highest abundances were Clostridia_vadinBB60_group, Bacteroides, Alistipes, Faecalibacterium, and Izemoplasmatales (Fig. 1C). On day 42, the phyla with the highest relative abundances were Firmicutes, Bacteroidota, Proteobacteria, Desulfobacterota, and Actinobacteriota (Fig. 2A). The most abundant families were Clostridia_vadin_BB60 group, Rikenellaceae, Ruminococcaceae, Bacteroidaceae, and Clostridia_UCG-014 (Fig. 2B). On day 42, the most abundant genera were Clostridia_vadinBB60_group, Alistipes, Bacteroides, Faecalibacterium, and Clostridia_UCG_014 (Fig. 2C). No significant differences in relative abundances were observed on either day of the experiment.

Relative abundance of phyla (A), families (B), and genera (C) in the cecal microbiota of 28-day-old broiler chickens. Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. Ctrl, control; C, vaccinated with coccidiosis only; CST, vaccinated with both coccidiosis and ST.

Relative abundance of phyla (A), families (B), and genera (C) in the cecal microbiota of 42-day-old broiler chickens. Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. Ctrl, control; C, vaccinated with coccidiosis only; CST, vaccinated with both coccidiosis and ST.
Analysis of alpha diversity revealed no significant differences between the groups on days 28 and 42 (Figs. 3 and 4). On day 28, beta diversity analysis based on Bray–Curtis distance (P = 0.024) and Jaccard distance (P = 0.023) showed significant differences when groups vaccinated against coccidiosis were compared to those that were not vaccinated (Fig. 5A). Beta diversity differences between the groups vaccinated against coccidiosis and those with no coccidia vaccination were also significant on day 42 (Bray–Curtis distance, P = 0.025; Jaccard distance, P = 0.028) (Fig. 5B). Vaccination against ST did not significantly influence beta diversity on day 28 (P = 0.495) (Fig. 6A) or 42 (P = 0.751) (Fig. 6B).

Alpha diversity measured by observed richness (A), Shannon diversity (B), and Simpson evenness (C) in the cecal microbiota of 28-day-old broiler chickens. Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. Ctrl, control; C, vaccinated with coccidiosis only; CST, vaccinated with both coccidiosis and ST.

Alpha diversity measured by observed richness (A), Shannon diversity (B), and Simpson evenness (C) in the cecal microbiota of 42-day-old broiler chickens. Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. Ctrl, control; C, vaccinated with coccidiosis only; CST, vaccinated with both coccidiosis and ST.

Beta diversity based on Bray–Curtis dissimilarity in the cecal microbiota of 28-day-old broilers (A) and 42-day-old broilers (B). Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. The groups vaccinated against coccidia are represented by the color red (groups C and CST) and the groups not vaccinated against coccidia are represented by the color green (groups Ctrl and ST). On both days, coccidiosis vaccination had a significant influence on the beta diversity (day 28: P = 0.024, day 42: P = 0.025). Ctrl, control.
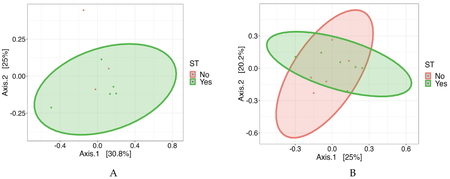
Beta diversity based on Bray–Curtis dissimilarity in the cecal microbiota of 28-day-old broilers (A) and 42-day-old broilers (B). Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. The groups vaccinated against ST are represented by the color green (groups ST and CST) and the groups not vaccinated against ST are represented by the color red (groups Ctrl and C). Ctrl, control.
In the functional pathway analysis, significant differences were observed between the treatment groups on day 28 (Fig. 7). Pathways associated with pentose and glucuronate interconversion (class: carbohydrate metabolism metabolites, P = 0.025), galactose metabolism (class: carbohydrate metabolism, P = 0.033), and arachidonic acid metabolism (class: lipid metabolism, P = 0.033) differed among the treatment groups. The abundance of these pathways was elevated in the group vaccinated against ST compared to that in other groups. Analysis of data from day 42 did not reveal any significant differences in functional pathways between the treatment groups.

Significantly altered functional pathways of cecal microbiota of 28-day-old broilers between all treatment groups. Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. Ctrl, control; C, vaccinated with coccidiosis only; CST, vaccinated with both coccidiosis and ST.
On day 28, four of the eight tested genes showed significant differences in expression (Fig. 8). Claudin 1 was overexpressed in group C compared to that in the control group (P = 0.042). Claudin 4 expression was higher in group C than in the control group (P = 0.012), as well as groups ST (P = 0.040) and CST (P < 0.001). E-cadherin was overexpressed in group C compared to that in group ST (P = 0.040). β-catenin was overexpressed in group C than in group Ctrl (P = 0.030). On day 42, 14 days after challenge with the ST marker strain, significant changes in the expression of two out of eight tight junction- related genes were observed (Fig. 8). Zonula occludens 2 was upregulated in group ST (P = 0.012) and in group C compared to that in the control group (P < 0.001). E-cadherin expression was significantly lower in group C than that in group ST (P = 0.031).

Relative gene expression of claudin 1 (CLDN1), claudin 2 (CLDN2), claudin 4 (CLDN4), claudin 5 (CLDN5), zonula occludens 1 (Zo1), zonula occludens 2 (Zo2), E-cadherin (Ecad), and β-catenin (Bcat) in the cecal wall of 28-day-old broilers. Birds were vaccinated against coccidiosis on day 1 (groups C and CST) and Salmonella Typhimurium (ST) on day 14 (groups ST and CST), and then challenged with the ST marker strain on day 28. Gene expression is normalized by the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hydroxymethylabilane synthase (HMBS) and expressed relative to the average of the uninfected birds. Letters indicate significant differences between groups (P ≤ 0.05). Ctrl, control; C, vaccinated with coccidiosis only; CST, vaccinated with both coccidiosis and ST.
The objective of this study was to assess how the efficacy of an ST vaccine was influenced by the administration of a coccidiosis vaccine and how the combination of both vaccines influenced intestinal health parameters. In poultry production, both vaccines are typically administered in the hatchery, with an ST booster administered on day 14. This pilot experiment was conducted to obtain proof-of-concept that both vaccines interacted with each other. For this reason, birds were vaccinated against ST only on day 14 because it seemed more likely that the potential influence of the coccidiosis vaccine on protection after ST vaccination would be more pronounced after coccidia had time to replicate. The administration route for the vaccine recommended by the manufacturer is via spray or drinking water[54]. However, oral gavage was selected as the route of administration in this study to ensure that each chicken received exactly one dose of the vaccine.
Previous studies have demonstrated a positive association between coccidiosis and salmonellosis in chickens[17,18,55]. In the current study, ST was isolated from cecal tonsils in all groups. However, ST was only detected in the livers of groups that received the coccidiosis vaccine. Consistent with the previously mentioned studies, these results indicate that the presence of coccidia infection enhances the establishment and systemic invasion of ST.
The reasons for this finding are unknown. To better understand the specific mechanism involved, the specific humoral immune response against Salmonella, cecal microbiota composition, and the expression of cell-junction genes in the ceca were investigated.
Given that ST primarily targets the intestinal tract, mucosal immunity, which is characterized by an IgA-dominated antibody response, serves as the first line of defense against microorganisms[56,57]. The current study revealed significantly decreased mean S/P value of IgA on day 42 in birds vaccinated against both coccidiosis and ST compared to those in all other groups. Additionally, there was a significant decrease in IgG levels in the cecal content of the group vaccinated against coccidiosis compared to that of the group vaccinated against ST. The reasons for these differences in antibodies are unclear.
Systemic antibodies play critical roles in targeting extracellular Salmonella[58]. Notably, no significant differences were observed in IgG antibody levels between the groups in either blood serum or cecal contents on day 42. This contrasts with the findings of Baba et al.[18], who have detected serum antibodies against S. Enteritidis (SE) only in birds co-challenged with coccidia and SE, but not in birds challenged solely with SE. We hypothesize that the vaccination before the challenge in the present study is the cause of this difference.
Analysis of microbiota and pathway data has confirmed that imbalances in the microbiota are associated with various disease states. The microbial composition on days 28 and 42 revealed no significant differences in the relative abundance of taxa or alpha diversity among treatment groups. Previous studies have also found no effect of coccidiosis on alpha diversity[59]. Research on the impact of ST vaccines on intestinal microbiota has observed no significant differences in relative abundance or alpha diversity compared to those in control groups[60]. However, one study reported elevated alpha diversity and significant differences in beta diversity in groups vaccinated against ST compared to those in the control group[61].
Beta diversity analysis based on Bray–Curtis and Jaccard distances showed significant differences between groups vaccinated with and without coccidiosis on days 28 and 42, indicating distinct microbial community structures in response to coccidiosis vaccination. Supplementation with coccidiostats, such as nicarbazin, does not affect microbiota composition in chickens or influence Salmonella invasion or colonization[62,63].
Significant differences in the functional KEGG Orthology pathways were observed among treatment groups on day 28. The pentose and glucuronate interconversions pathway plays a crucial role in detoxification and metabolite excretion, potentially contributing to reduced oxidative stress[64,65]. As a precursor to proinflammatory mediators, arachidonic acid significantly influences inflammation, while its metabolites are closely linked to oxidative stress, lipid metabolism, and immune function[66,67,68]. The high abundance of sequences associated with these metabolic pathways suggests increased inflammation response in the chicken cecum, when chicken are vaccinated and challenged with ST.
Overall, these findings highlighted the influence of both vaccinations on the cecal microbial community and its metabolic activities.
One of the key components of the intestinal barrier is the formation of tight junctions, adherens junctions, gap junctions, and desmosomes between epithelial cells[69,70]. These junctions, which are multiprotein complexes, seal the paracellular space between adjacent epithelial cells and regulate intestinal barrier permeability[71,72]. Pathogen-induced damage to these junction complexes and disruption of the intestinal barrier allow macromolecules, such as antigens, bacterial toxins, and pathogens from the intestinal lumen, to cross into circulation[73].
Our findings revealed changes in the expression of claudin 1 on day 28, claudin 4 on days 28 and 42, and zonula occludens 2 on day 42. Claudins are primary transmembrane proteins that contribute to tight junctions, while zonula occludens 1 serves as a cytoplasmic plaque protein that interacts with both transmembrane and cytoskeletal proteins[74,75]. Salmonella infection downregulates claudin 1 gene expression in the ileum and jejunum of broiler chickens[76]. Similarly, ST infection reduces claudin 1 and claudin 4 expression in the jejunum of broiler chickens[77].
E-cadherin was differentially expressed on day 28. This gene is involved in growth regulation and cell-cell adhesion in enterocytes, with increased levels observed in proliferating cells[78]. β-catenin is involved in a pathway responsible for intestinal cell growth[79]. Previous studies have shown elevated β-catenin levels following Salmonella infection in chickens[78,80]. Overall, more significant differences were observed between groups before the ST challenge (day 28), possibly because the effect of the ST challenge was greater than the subtle differences between vaccination programs.
This pilot experiment indicated that coccidiosis vaccination interfered with the efficacy of ST vaccination administered on day 14, although the results were not significantly conclusive. Minor differences were also observed in the intestinal antibody response, microbiota abundance, and expression of tight junction genes. Larger-scale studies are required to fully investigate whether coccidiosis vaccination influences the effectiveness of ST vaccination following the recommended schedule on days 1 and 14.
This research was partially funded by the United States Department of Agriculture-Agricultural Research Services (USDA-ARS, Project Number: 6040-32000-085-002-S) and the Alabama Agricultural Experiment Station.
Conceptualization: Rüdiger Hauck, Kenneth Macklin, and Xu Wang; formal analysis, Rüdiger Hauck, Andrea Pietruska; investigation, Rüdiger Hauck, Andrea Pietruska, Teresa Dormitorio, James Krehling; data curation, Andrea Pietruska; writing—original draft preparation, Andrea Pietruska; writing—review and editing, Andrea Pietruska, Rüdiger Hauck; project administration, Rüdiger Hauck; funding acquisition, Rüdiger Hauck, Kenneth Macklin and Xu Wang All authors have read and agreed to the published version of the manuscript.
The authors declare no conflict of interest.