2012 年 62 巻 4 号 p. 334-339
2012 年 62 巻 4 号 p. 334-339
It has long been known that a bacterial leaf blight-resistant line in rice obtained from a crossing using ‘Asominori’ as a resistant parent also has resistance to blast, but a blast resistance gene in ‘Asominori’ has not been investigated in detail. In the present study, a blast resistance gene in ‘Asominori’, tentatively named Pias(t), was revealed to be located within 162-kb region between DNA markers YX4-3 and NX4-1 on chromosome 4 and to be linked with an ‘Asominori’ allele of the bacterial leaf blight resistance gene Xa1, tentatively named Xa1-as(t). An ‘Asominori’ allele of Pias(t) was found to be dominant and difference of disease severity between lines having the ‘Asominori’ allele of Pias(t) and those without it was 1.2 in disease index from 0 to 10. Pias(t) was also closely linked with the Ph gene controlling phenol reaction, suggesting the possibility of successful selection of blast resistance using the phenol reaction. Since blast-resistant commercial cultivars have been developed using ‘Asominori’ as a parent, Pias(t) is considered to be a useful gene in rice breeding for blast resistance.
Rice blast caused by Magnaporthe grisea is a major disease in rice, Oryza sativa L. and a large amount of fungicide is used to reduce yield loss due to an epidemic of blast disease in Japan. Especially in the Tohoku region, northeastern area in Japan, low temperature and a low level of sunshine are frequent in summer, causing not only sterility but also an epidemic of blast, resulting in a severe loss of yield. Therefore, cold tolerance and blast resistance are the most important breeding objectives for rice in this region. Although an epidemic of bacterial leaf blight caused by Xanthomonas oryzae became less frequent in Japan, because occurrence of overhead flooding was reduced by civil engineering of paddy fields, it is still an important disease in Asia.
Blast resistance is controlled by several major genes for race-specific true resistance and many minor genes for race-nonspecific field resistance (Asaga 1981). Since blast resistance by a single gene for true resistance has been broken down by a new compatible race (Kiyosawa 1982) and cultivation of multiple lines having different true resistance genes as a mixture has been demonstrated to be effective for preventing breakdown of true resistance (Koizumi et al. 2004, Mundt 2002, Zhu et al. 2000), several isogenic lines having different true resistance genes, i.e., multilines, have been developed by repeated backcrossing of ‘Sasanishiki’ or ‘Koshihikari’ as a recurrent parent (Abe 2004, Ishizaki et al. 2005). Field resistance, on the other hand, is stable and durable with little possibility of a breakdown of resistance; therefore, stacking of field resistance genes from different Japanese cultivars and breeding using powerful field resistance derived from upland rice cultivars have been carried out in Japan (Higashi 1995, Saka 2006, Yamaguchi 2009). As field resistance genes, Pb1 (Fujii et al. 1999, Hayashi et al. 2010), pi21 (Fukuoka and Okuno 2001), Pi39(t) (Terashima et al. 2008), Pi34 (Zenbayashi et al. 2007) and Pi35 (Nguyen et al. 2006) have been reported and blast resistant cultivars have been developed using these genes, e.g., ‘Koshihikari-AichiSBL’, ‘Tomohonami’, ‘Mineharuka’, ‘ChugokuIL1’, ‘ChugokuIL2’ and ‘Yukinohana’. Among these field resistance genes, Pb1 exhibits resistance to panicle blast. A DNA marker closely linked with Pb1 has been developed, and utility of this DNA marker in rice breeding has been demonstrated (Sugiura et al. 2004). As a result, ‘Tsukinohikari’, ‘Matsuribare’,’Asahinoyume’ and ‘Daichinokaze’ carrying Pb1 have been bred. Recently, pi21 derived from an upland rice cultivar has been revealed to be linked with a gene for eating quality and is expected to be useful in rice breeding (Fukuoka et al. 2009).
‘Asominori’ developed in 1973 by a cooperative breeding program between Kumamoto Prefecture and Kyushu Agricultural Research Station (NARO Kyushu Okinawa Agricultural Research Center at present) has a high level of field resistance to bacterial leaf blight (Sugitani et al. 1976). Xa1-as(t) at the Xa1 locus is a dominant allele controlling resistance to three isolates, T7174 (race I), H9153 (race II-2) and H75304 (race V) of bacterial leaf blight in ‘Asominori’ (Ise 1998). Xa1-as(t) provides resistance to race I and race V of Xanthomonas oryzae at both the young seedling stage and the adult stage, and is therefore considered to be different from Xa1, which causes unstable resistance at the young seedling stage. Disease resistance of ‘Asakaze’ and ‘Kougyoku’ having Xa1 has been broken down, but ‘Asominori’ having Xa1-as(t) has been resistant for more than 35 years. Xa1-as(t) has been reported to be closely linked, the recombination rate being 2.4%, with Ph for the phenol reaction, which turns rice seeds soaked in phenol solution dark brown (Ise 1998). It has been empirically known by some Japanese rice breeders that not only bacterial blight resistance but also blast resistance can be acquired by using ‘Asominori’ as a resistant parent. Closely linked genes for blast and bacterial blight are useful for efficient development of disease resistant cultivars in rice. However, the presence of a blast resistance gene in ‘Asominori’ has not been confirmed.
Recently, analyzing backcrossed progeny between ‘Asominori’ as a non-recurrent parent and ‘Okiniiri’ as a recurrent parent, we have revealed a blast resistance gene and Xa1-as(t) in ‘Asominori’ to be within 1336-kb on chromosome 4 (Endo et al. 2009), but have not identified the loci of these genes. In the present study, we newly obtained recombinants in this region and clarified the relation between these resistance genes to be a linkage. Then, we tentatively named the blast resistance gene in ‘Asominori’ Pias(t) and localized it near Xa1-as(t) on chromosome 4.
Nineteen lines obtained by crossing of ‘Hitomebore’, which is susceptible to blast and bacterial leaf blight and negative in phenol reaction, with a progeny of a cross between ‘Asominori’, resistant to blast and bacterial leaf blight and positive in phenol reaction, and ‘Okiniiri’, which is less resistant to blast, susceptible to bacterial leaf blight and negative in phenol reaction, were used as materials. The pedigree of the employed materials and a genealogy of Asominori are shown in Supplemental Figs. 1, 2, respectively. ‘Okiniiri’ was crossed with ‘Asominori’ in 1997 and an F1 plant was backcrossed with ‘Okiniiri’. By testing field resistance to leaf blast, a blast-resistant line, ‘Ukei854’ (later named ‘Ouu 411’), was selected. An F3 plant derived from a cross between ‘Ukei854’ and ‘Hitomebore’ was crossed with ‘Hitomebore’ to obtain an F1 plant. From 826 F2 plants obtained by selfing of this F1 plant, two recombinants between YX4-3 and NX4-1 were selected and four and three lines of group 1 and group 2, respectively, were developed. Similarly, three homozygotes of ‘Hitomebore’ or ‘Asominori’ alleles between RM5511 and RN5709 were selected from the 826 F2 plants and three lines of group 4 and group 5 each were developed. From 20 B1F1 plants obtained by backcrossing of ‘Hitomebore’, one recombinant of group 3 was selected, and three recombinant inbred lines were developed. Seeds of ‘Saikai 85’, ‘Saikai 59’, ‘Sachikaze’, ‘Pi No. 2’, ‘Jukkoku’, ‘Zenshou 26’, ‘Tadukan’ and ‘Senbonasahi’ were obtained from the NIAS genebank.
DNA polymorphism analysisDNA was extracted from fresh leaves using TPS solution (100 mM Tris-HCl, pH 8.0, 10 mM ethylendiaminetetra-acetic acid, 1 M KCl) according to the method of Thompson and Henry (1995). DNA was precipitated by the addition of isopropanol, washed with ethanol and dissolved in 1/10 TE to be used as a PCR template. The PCR condition was 7 min at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 50–65°C and 2 min at 72°C and a final extension for 10 min at 72°C. PCR products were electrophoresed in 3% agarose gel. CAPS markers were analyzed after digestion of PCR products at 37°C overnight with the restriction endonucleases shown in Supplemental Table 1. DNA markers used in the present study are listed in Supplemental Table 1. Primer sequences of “RM” markers were obtained from Gramene (http://www.gramene.org/) and primers of the other markers were designed using the genome sequence of Nipponbare (http://rgp.dna.affrc.go.jp/IRGSP/).
Phenol reactionThree seeds per line were soaked in 1.5% phenol solution for 3 hours and dried for observing seed color. Black-colored seeds were scored as positive.
Disease resistanceBlast resistance of the lines was evaluated by the upland nursery test with natural infection at the NARO Tohoku Agricultural Research Center, Daisen, Akita, Japan in 2008 and 2009 and at Miyagi Prefectural Furukawa Agricultural Experiment Station, Osaki, Miyagi, Japan in 2010. Approximately 100 seeds per line were sown in a row 20–35 cm long having 8–10 cm spacing early in June with duplication in 2008 and with triplication in 2009 and 2010. A susceptible cultivar, ‘Inabawase’ and a susceptible line, ‘54kei3110’, were planted at the side of plot area to enhance the natural infection in 2008 and 2009 and in 2010, respectively. Disease severity was scored 0 (no symptom) to 10 (death of whole leaves) according to Asaga (1981) three times from late July to early August in 2008–2010.
Resistance to bacterial leaf blight was examined by an inoculation test using the leaf clipping method (Ogawa and Sekizawa 1980). Plants were grown with basal fertilizer of 70 kgN/ha and three top dressings of 20 kgN/ha to enhance susceptibility. One week before heading, flag leaves were inoculated by cutting with scissors drenched with bacterial suspension of strain T7174 (race I). Two to three weeks after inoculation, disease severity was rated with a length of chlorosis in flag leaves.
Graphical genotyping data of the tested lines are shown in Fig. 1. Based on genotypes of DNA markers between RM5511 and RM5709, the lines were classified into five groups. The number of lines in group 1 was four and in each of the other groups was three. In group 1 to group 3, recombination has occurred between RM5511 and RM5709. Group 4 and group 5 were homozygous for ‘Hitomebore’ alleles and ‘Asominori’ alleles, respectively, at all the loci between RM5511 and RM5709. ‘Okiniiri’, a recurrent parent of ‘Ouu 411’, belonged to group 4 and ‘Ouu 411’ belonged to group 5. Physical distance between RM5511 and RM5709 is ca. 948 kb in ‘Nipponbare’ (http://rgp.dna.affrc.go.jp/IRGSP/).
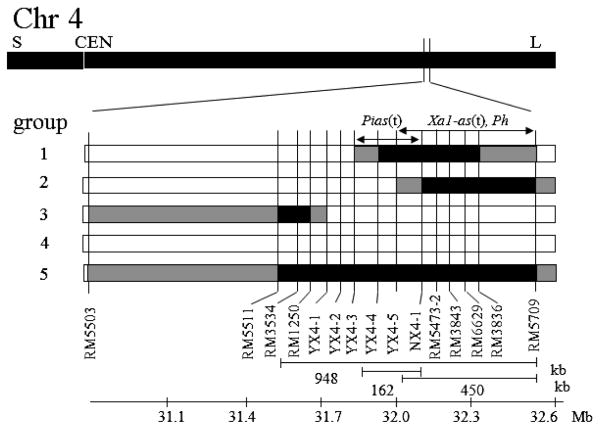
Loci of Pias(t), Xa1-as(t) and Ph on chromosome 4. Black and white columns represent ‘Asominori’ and ‘Hitomebore’ alleles, respectively. Gray columns indicate regions containing recombinant break points. Physical distances between DNA markers were estimated from the genome sequence of ‘Nipponbare’.
Disease severity of blast of group 4 lines was 5.0 on average of two years, which is similar to that of ‘Hitomebore’, and that of group 5 lines was 3.8, which is comparable to those of ‘Asominori’ and ‘Ouu 411’ (Fig. 2). Disease severity of group 1 lines was 3.7 and those of group 2 lines and group 3 lines were 5.1 and 4.7, respectively. The values of group 1 and 5 were significantly lower than that of group 4 at the 1% level by Student’s t-test. The data at Daisen in 2009 corresponded well to those at Osaki in 2010. Group 1 having ‘Asominori’ alleles of the markers between YX4-4 to RM3836 was resistant, while group 2 having ‘Asominori’ alleles of the markers between NX4-1 and RM5709 was susceptible, suggesting that a gene for blast resistance is located within a ca. 162-kb region between YX4-3 and NX4-1. Based on the difference of disease severity scores between group 4 and group 5, substitution of this region from homozygous ‘Hitomebore’ genotype to homozygous ‘Asominori’ genotype is estimated to reduce the disease severity score of 1.2. In the examination of disease resistance using an F3 population in 2008, ‘Hitomebore’-type homozygous lines (n = 4), ‘Asominori’-type homozygous lines (n = 2) and heterozygous lines (n = 10) in the region from RM3534 to RM3836 exhibited disease severity scores of 7.8, 4.3 and 5.0, respectively. Since the value of the heterozygous lines was similar to that of the ‘Asominori’-type homozygous lines and significantly different (P < 0.01) from that of the ‘Hitomebore’-type homozygous lines, the ‘Asominori’-type was considered to be dominant over the ‘Hitomebore’-type.
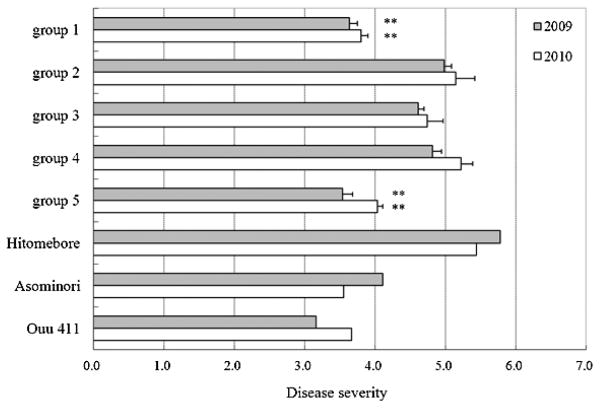
Disease severity of rice blast in breeding lines. Gray and white columns represent disease severity of rice blast scored at Daisen in 2009 and Osaki in 2010, respectively. The error bars show standard error. Double asterisks indicate 1% significant difference from disease severity of group 4. Score of each group was an average of three time scoring with three repetitions of three lines (group 2–5) or four lines (group 1).
In the inoculation test of bacterial leaf blight, being susceptible, ‘Hitomebore’ and lines of group 3 and 4 showed extension of chlorosis, while, being resistant, ‘Asominori’, ‘Ouu 411’ and lines of group 1, 2 and 5 showed necrosis at the inoculated leaf ends but no extension of chlorosis (Table 1). ‘Hitomebore’ and the lines of group 3 and 4 were negative to phenol reaction, but ‘Asominori’, ‘Ouu 411’ and the lines of group 1, 2 and 5 showed a positive reaction. These results suggest that Xa1-as(t) and Ph are located in the ca. 450-kb region between YX4-5 and RM5709. Segregation of blast resistance and bacterial leaf blight resistance in the progeny derived from ‘Asominori’ indicates that a gene for blast resistance is different from but linked with Xa1-as(t). Therefore, the blast resistance gene of ‘Asominori’ was tentatively named Pias(t).
| Trait | Group No. or cultivar | 1 | 2 | 3 | 4 | 5 | Hitomebore | Asominori | Ouu411 |
|---|---|---|---|---|---|---|---|---|---|
| BLB resistancea | R | R | S | S | R | S | R | R | |
| Phb | + | + | − | − | + | − | + | + |
Disease resistance, phenol reaction and genotypes of DNA markers from YX4-3 to RM5709 in ancestors of ‘Asominori’ are shown in Table 2. ‘Saikai 85’, ‘Pi No. 2’ and ‘Tadukan’ were resistant to rice blast, disease severity being 1.0, 1.7 and 0.0, respectively. It has already been found that ‘Saikai 85’, ‘Pi No. 2’ and ‘Tadukan’, but not ‘Asominori’, carried Pita, which is another resistance gene to rice blast (Hayashi et al. 1998, http://ineweb.narcc.affrc.go.jp/index.html). ‘Saikai59’, ‘Sachikaze’, ‘Jukkoku’, ‘Zenshou 26’ and ‘Senbonasahi’ were susceptible to rice blast, while ‘Saikai 85’ and ‘Tadukan’ were resistant to bacterial leaf blight. ‘Zenshou 26’ was assessed to be susceptible to bacterial leaf blight, although it is known as a cultivar resistant to bacterial leaf blight (Noda and Ohuchi 1989). In phenol reaction, only ‘Saikai 85’ was positive. Genotypes of DNA markers from YX4-3 to RM5709 in ‘Saikai 85’ were ‘Asominori’-type. ‘Tadukan’ was ‘Asominori’-type except YX4-4, NX4-1, RM6629 and RM5709. On the other hand, genotypes in these loci of ‘Saikai 59’, ‘Sachikaze’, ‘Pi No. 2’ were ‘Hitomebore’-type.
| Trait or Markerd | Saikai85 | Saikai59 | Sachikaze | Pi No. 2 | Jukkoku | Zenshou 26 | Tadukan | Senbonasahi |
|---|---|---|---|---|---|---|---|---|
| LB resistancea | 1.0 | 4.3 | 5.3 | 1.7 | 5.2 | 5 | 0 | 3.8 |
| BLB resistanceb | R | S | S | S | S | S | R | S |
| Phc | + | − | − | − | − | − | − | − |
| YX4-3 | A | H | H | H | H | H | A | H |
| YX4-4 | A | H | H | H | H | H | H | H |
| YX4-5 | A | H | H | H | H | H | A | H |
| NX4-1 | A | H | H | H | H | H | – | H |
| RM5473-2 | A | H | H | H | H | H | A | H |
| RM3843 | A | H | H | H | H | H | A | H |
| RM6629 | A | H | H | H | H | H | C | H |
| RM3836 | A | H | H | H | H | H | A | H |
| RM5709 | – | H | H | H | H | H | – | H |
–: not amplified.
Agronomic traits of ‘Ouu 411’ having Pias(t) and Xa1-as(t) are shown in Supplemental Table 2. Although the number of panicles of ‘Ouu 411’ was less than that of ‘Hitomebore’ and ‘Akitakomachi’, which are leading cultivars in the Tohoku region of Japan, grain weight was heavier in ‘Ouu 411’ than in ‘Hitomebore’ and ‘Akitakomachi’, resulting in comparable yields among them. Both resistances to leaf blast and panicle blast were estimated to be level 3, i.e., high resistance.
As blast resistance genes which provide a high level of field resistance, pi21 (Fukuoka et al. 2009), Pb1 (Hayashi et al. 2010), Pi9 (Qu et al. 2006), Pi34 (Zenbayashi et al. 2007), Pi35 (Nguyen et al. 2006), Pi40 (Jeung et al. 2007), Pikahei-1(t) (Miyamoto et al. 2001, Xu et al. 2008), Pi39(t) (Terashima et al. 2008) and so on have been reported. Among them, Pikahei-1(t) and Pi39(t) have been mapped on the long arm of chromosome 4, where Pias(t) was found to be located. Disease severity of ‘Ouu 411’ having Pias(t) was 1.8, while those of ‘Mineharuka’ having Pi39(t) and ‘Kahei’ having Pikahei-1(t) were 1.5 and 1.0, respectively. Pikahei-1(t) has been found to be located in the 300 kb region between RM17496 and RM6629. Pias(t) was located in the 162 kb region included in this 300 kb region. ‘Kahei’ is susceptible to bacterial leaf blight of strain T7174 (race I) and is a local variety of Japanese upland rice. Pi39(t), which is derived from a Chinese local variety, is also located in the region near Pias(t). Since the phylogenetic relationship between ‘Asominori, ‘Kahei’ and the Chinese local variety is unknown, the possiblity that Pias(t) is the same as Pikahei-1(t) or Pi39(t) cannot be ruled out. Only ‘Saikai 85’ among the parental lines of ‘Asominori’ had the same genotypes of the DNA markers as those of ‘Asominori’, suggesting ‘Saikai 85’ to be the gene source of Pias(t). However, all the parental lines of ‘Saikai 85’, i.e., ‘PiNo. 2’, ‘Jukkoku’ and ‘Zenshou 26’, were susceptible to bacterial leaf blight and had ‘Hitomebore’ alleles of all the DNA markers between YX4-3 and RM5709. The original gene source of Pias(t) was not identified in the present study.
Xa1 in ‘IR24’ has been reported to be a disease resistance gene having the NBS-LRR structure (Yoshimura et al. 1998). Exhaustive analysis of disease resistance genes around Xa1 on the long arm of chromosome 4 has revealed this region to be rich in NBS-LRR sequences (Monosi et al. 2004). The presence of several NBS-LRR sequences in a region near Xa1 made it difficult to identify a candidate gene for Pias(t). In the present study, a region containing Pias(t) was delimited to a ‘Nipponbare’ genome sequence of ca. 162 kb by fine mapping using gene-derived DNA markers. Since several genes related to disease resistance are annotated in this region by RAP-DB (http://rapdb.dna.affrc.go.jp), Pias(t) is expected to be identified at the molecular level by analysis of these genes.
Since ‘Miyako95’ (Nishiyama et al. 1992), ‘Yokaminori’ and ‘Nankai 119’, which are resistant to both blast and bacterial leaf blight, have been developed from progeny of ‘Asominori’, Pias(t) and Xa1-as(t) are considered to be unlinked with any genes for fatal unfavorable characteristics. In fact, ‘Ouu 411’ developed in the present study has characteristics comparable to leading cultivars (Supplemental Table 2). Since Pias(t) and Xa1-as(t) are closely located within a 612-kb region, this region is inferred to be highly useful in rice breeding for disease resistance. Furthermore, selection of lines resistant to both blast and bacterial leaf blight may be achieved using the phenol reaction in addition to the DNA markers developed in the present study.
Although many blast resistance genes have been reported (Koide et al. 2009, Xu et al. 2004), there have been few studies evaluating the effects of field resistance genes introduced into commercial varieties on suppression of disease symptom development. It is important to assess the effects of blast resistance genes in commercial varieties. Use of various disease resistance genes and stacking various blast resistance genes in a cultivar are considered to be effective for developing a stable blast-resistant cultivar. Further studies for identification of loci of many blast resistance genes and development of DNA markers will contribute to the development of rice cultivars having stable resistance to blast.
The pathogen stock of bacterial leaf blight was provided by NARO Kyushu Okinawa Agricultural Research Center. DNA polymorphism analysis was carried out at the laboratory of the QTL Genome Center at the National Institute of Agrobiological Sciences.