2012 年 62 巻 4 号 p. 348-351
2012 年 62 巻 4 号 p. 348-351
Japanese pear (Pyrus pyrifolia) exhibits gametophytic self-incompatibility (GSI) controlled by a complex and multiallelic S locus. The pistil-part product of the S locus is the polymorphic ribonuclease, S-RNase. Information on S-genotypes is important for the production and breeding of Japanese pears. Molecular analyses of S-genotypes of Japanese pear have been conducted with the CAPS (cleaved amplified polymorphic sequence) system; PCR amplification of S-RNase fragments by a common primer pair followed by digestion with restriction enzymes each of which cleaves a specific S haplotype. Here, we show that the separation of S-RNase fragments by polyacrylamide gel electrophoresis (PAGE) distinguishes four out of nine S haplotypes of Japanese pear without restriction digestion. S3-, S5-, S6- and S8-RNases were identified as distinct bands by PAGE. S3- and S5-RNases were separated by PAGE despite their identical fragment sizes. Using this system, three Japanese pear lines with unknown S-genotypes were analyzed. The newly determined S-genotypes of the lines were confirmed by CAPS analysis.
Many fruit trees of Rosaceae such as Japanese pear, apple and sweet cherry exhibit gametophytic self-incompatibility (GSI) in which a polymorphic S locus is involved in self/non-self discrimination. When an S haplotype of pollen matches one of the two S haplotypes of a pistil, the pollen is recognized as self and rejected (de Nettancourt 2001, Franklin-Tong 2008). For self-incompatible fruit trees, artificial pollination with compatible pollen or the planting of compatible pollen donor plants is necessary to guarantee stable fruit production. Therefore, information on S-genotypes of cultivars is important for selecting appropriate pollen donors. For the breeding of fruit trees with GSI, information on S-genotypes is also necessary for the selection of appropriate cross combinations. S-genotypes have been analyzed by pollination studies in Japanese pear (Pyrus pyrifolia) (Machida et al. 1982, Terami et al. 1946). However, pollination tests are heavily affected by environmental conditions and can only be conducted in the flowering season.
Pistil-part S specificity is controlled by a highly polymorphic ribonuclease (RNase) called S-RNase in rosaceous species (Sassa et al. 1992, 1996, 1997, Tao et al. 1997, 1999, Ushijima et al. 1998). Consequently, analyses of sequence polymorphisms of the S-RNase gene have been used to determine S-genotypes of rosaceous fruit trees. The S-RNase-based genotyping methods are divided into two types: specific primer-based methods using different PCR primer sets each of which amplifies a specific S-RNase allele only and CAPS (cleaved amplified polymorphic DNA) or PCR-RFLP analyses, which amplify S-RNase fragments via a common primer set designed to amplify all the S-RNase alleles and then treat the products with restriction enzymes each of which digests a specific allele to detect S alleles as specific restriction fragments. The specific primer-based methods, however, are sometimes hampered by false-negative and false-positive problems. Poor quality and/or quantity of template DNA result in false negatives. Some primer pairs were even reported to amplify non-target alleles (Ershadi and Talaii 2007, Matsumoto et al. 2000). Although the CAPS system is thought to cope with these false-negative/positive problems, it requires expensive restriction enzyme sets and extra time for the digestion of PCR products. While S-RNase is highly polymorphic, distinguishing some exceptionally related pairs is problematic for CAPS. S3- and S5-RNases of Japanese pear are highly homologous (97% nucleotide identity) and an S3-RNase-specific restriction enzyme is not available. S3-RNase is therefore detected as a PCR product that is digested by PpuMI, which cleaves both S3- and S5-RNases, but not cut by the S5-specific enzyme AlwNI (Ishimizu et al. 1999).
In this paper we report that the polyacrylamide gel electrophoresis (PAGE), instead of the widely used agarose gel electrophoresis (AGE), of PCR-amplified S-RNase fragments is useful for S-genotyping in Japanese pear. Among the nine S-RNase fragments amplified by a common primer pair for CAPS, four alleles were distinguished by PAGE without restriction treatment. It is worth noting that very similar alleles, S3 and S5, were also distinguishable by PAGE having PCR products of the exact same length.
Twelve Japanese pear cultivars were used, Doitsu (S1S2), Chojuro (S2S3), Natsuhikari (S3S4), Kosui (S4S5), Shinsetsu (S5S6), Imamura-aki (S1S6), Hogetsu (S1S7), Okusankichi (S5S7), Ichiharawase (S1S8), Niitaka (S3S9), Shinko (S4S9) and Hosui (S3S5). Homozygous plants S1S1 and S6S6 were produced by self pollination of young flower bud of Imamura-aki (S1S6). S3S3 and S4S4 plants were produced by self pollination of Natsuhikari (S3S4). Genotypes of these homozygous plants were determined by CAPS analysis (Ishimizu et al. 1999) (Kato and Sassa, unpublished results). 3-W26-12 is a progeny of Wakahikari (S3S4) × 26-12 (S3S9). 2-WA-12 and 2-WA-26 are progenies of Wakahikari (S3S4) × Aikansui (S4S5).
DNA extraction, PCR and CAPSDNA was extracted from leaves (Sassa 2007) and used for PCR amplification of S-RNase fragments with the primer pair FTQQYQ and anti-(I/T)IWPNV as described (Ishimizu et al. 1999, Takasaki et al. 2004). Length of the PCR products of S1-, S2-, S3-, S4-, S5-, S6-, S7-, S8- and S9-RNases are 367, 1347, 376, 368, 376, 347, 352, 436 and 1307 bp, respectively (Ishimizu et al. 1999, Takasaki et al. 2004). A genomic clone of S5-RNase was derived from the PCR product amplified from leaf DNA of Kosui (S4S5) by primer 7 and primer 5 (Sassa et al. 1997) and used as a template for PCR amplification of the S5-RNase fragment. CAPS was conducted as described (Ishimizu et al. 1999, Takasaki et al. 2004).
PAGEAmplified S-RNase fragments were separated on an 8% polyacrylamide gel with a discontinuous buffer system (Davis 1964) essentially as described (Kawaguchi et al. 2001). Briefly, composition of separation gel is 0.375 M Tris-HCl pH 8.8, 8% acrylamide and 0.21% N,N′-methylenebisacrylamide. Stacking gel contains 0.13 M Tris-HCl pH 6.8, 5% acrylamide and 0.13% N,N′-methylenebisacrylamide. Electrophoresis was conducted with the running buffer containing 0.25 M Tris and 1.92 M glycine. After the electrophoresis, the gel was stained with ethidium bromide.
S-RNase fragments were amplified from eleven Japanese pear cultivars with known S-genotypes, four S homozygous plants (S1S1, S3S3, S4S4 and S6S6) and a genomic clone of S5-RNase and separated by AGE and PAGE (Fig. 1). AGE detected two bands; a short band corresponding to S1-, S3-, S4-, S5-, S6-, S7- and S8-RNases and a long band corresponding to S2- and S9-RNases as described before (Ishimizu et al. 1999, Takasaki et al. 2004) (Fig. 1A). On the other hand, separation of the S-RNase fragments by PAGE enabled the discrimination of four S-RNases detected as short bands in AGE (Fig. 1B). Although S1-, S4- and S7-RNases were not separated, S3-, S5-, S6- and S8-RNases were identified as distinct bands. It is worth noting that S3- and S5-RNases were distinguishable by PAGE, although these fragments are identical in size (376 bp; Ishimizu et al. 1999, Takasaki et al. 2004). Furthermore, because S3- and S5-RNases are very similar (97% nucleotide identity; Ishimizu et al. 1998), an S3-specific restriction enzyme is not available for S-genotyping by CAPS (Ishimizu et al. 1999). S3-RNase is detected as a band cleaved by PpuMI that digests both S3- and S5-RNases, but is not cut by the S5- specific enzyme AlwNI. Although why PAGE can separate S3- and S5-RNases of the same length is not clear, Tsubokura et al. (2006) also detected sequence variations of PCR fragments with identical sizes as polymorphic bands on PAGE. Ishimizu et al. (1999) analyzed the PCR products of S-RNases of Japanese pear by PAGE and showed that some S-alleles can be distinguished without restriction treatment. However, S3- and S5-RNases comigrated as a single band and could not be distinguished (Ishimizu et al. 1999). This may be because the buffer system used for PAGE was different from our discontinuous system (Davis 1964, Kawaguchi et al. 2001), although the experimental details for PAGE are not described in Ishimizu et al. (1999).

Separation of the S-RNase fragments of Japanese pear by aga-rose and polyacrylamide gel electrophoresis. (A) AGE (1% agarose in TAE buffer). (B) PAGE. The lanes of each gel; 1, S1S1; 2, S3S3; 3, S4S4; 4, S5 clone; 5, S6S6; 6, Doitsu (S1S2); 7, Chojuro (S2S3); 8, Natsuhikari (S3S4); 9, Kosui (S4S5); 10, Shinsetsu (S5S6); 11, Imamura-aki (S1S6); 12, Hogetsu (S1S7); 13, Okusankichi (S5S7); 14, Ichiharawase (S1S8); 15, Niitaka (S3S9); 16, Shinko (S4S9).
By using the PAGE analysis of S-RNases, S-genotypes of three Japanese pear lines were analyzed. In the breeding program at the Chiba Prefectural Agriculture and Forestry Research Center, 3-W26-12 was selected from the progenies of Wakahikari (S3S4) × 26-12 (S3S9). 2-WA-12 and 2-WA-26 are progenies of Wakahikari (S3S4) × Aikansui (S4S5). Therefore, the S-genotype of 3-W26-12 is expected to be either S3S9 or S4S9 as S3 pollen from 26-12 (S3S9) is rejected by the pistil of Wakahikari (S3S4). Similarly, 2-WA-12 and 2-WA-26 are expected to be either S3S5 or S4S5 because S4 pollen from Aikansui (S4S5) is rejected by the pistil of Wakahikari (S3S4). Consistent with this, 2-WA-26 showed both long and short bands while 2-WA-12 and 2-WA-26 showed short bands only on AGE (Fig. 2A). PAGE of the S-RNase fragments showed that 3-W26-12 has the S3 haplotype and thus is expected to be S3S9, and the S-genotypes of both 2-WA-12 and 2-WA-26 are S4S5 (Fig. 2B).

Analysis of S-RNases for S genotyping of three lines, 3-W26-12, 2-WA-12 and 2-WA-26. (A) AGE. (B) PAGE.
To confirm the S-genotypes of the three lines, CAPS analysis of S-RNases (Ishimizu et al. 1999, Takasaki et al. 2004) was conducted. The ~400 bp fragment of 3-W26-12 was digested by the S3- and S5-specific enzyme PpuMI but not by S5-specific enzyme AlwNI, indicating that the fragment corresponds to S3-RNase (Fig. 3A, 3B). The ~1.4 kbp band of 3-W26-12 was digested by BstBI, showing that it was derived from S9-RNase (Fig. 3C). Digestion of the PCR products of 2-WA-12 and 2-WA-26 with the S5-pecific enzyme AlwNI and S4-pecific enzyme NdeI further showed that the S-genotypes of 2-WA-12 and 2-WA-26 are S4S5 (Fig. 3B, 3D).
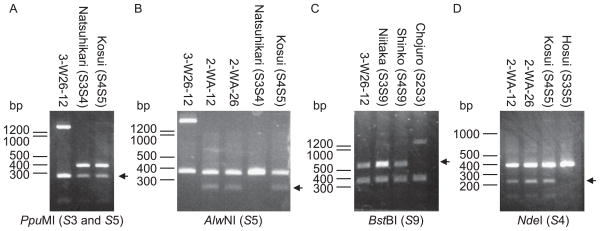
CAPS analysis of S-RNases for S genotyping of 3-W26-12, 2-WA-12 and 2-WA-26. PCR products were digested by S haplotype-specific restriction enzymes before AGE; (A) PpuMI for S3 and S5, (B) AlwNI for S5, (C) BstBI for S9 and (D) NdeI for S4. The S haplotype-specific restriction enzymes used are indicated under the AGE images. Arrows denote respective S haplotype-specific restriction fragments.
This study revealed that PAGE analysis of S-RNase fragments enables the discrimination of four out of nine S haplo-types without restriction enzymes and thus is a simple and effective means for S-genotyping of Japanese pear. This method would also be useful for genotyping other rosaceous fruit trees such as apple and European pear in which common primer pairs for CAPS systems have been established (Kim et al. 2009, Moriya et al. 2007).
This work was supported by the Grants-in-Aid for Scientific Research (B, 24380004) from the Ministry of Education, Science, Sports and Culture of Japan.