2019 Volume 44 Issue 11 Pages 721-735
2019 Volume 44 Issue 11 Pages 721-735
For the safety assessment of pharmaceuticals, initial data management requires accurate toxicological data acquisition, which is based on regulatory safety studies according to guidelines, and computational systems have been developed under the application of Good Laboratory Practice (GLP). In addition to these regulatory toxicology studies, investigative toxicological study data for the selection of lead compound and candidate compound for clinical trials are directed to estimation by computational systems such as Quantitative Structure-Activity Relationship (QSAR) and related expert systems. Furthermore, in the “Go” or “No-Go” decision of drug development, supportive utilization of a scientifically interpretable computational toxicology system is required for human safety evaluation. A pharmaceutical safety evaluator as a related toxicologist who is facing practical decision needs not only a data-driven Artificial Intelligence (AI) system that calls for the final consequence but also an explainable AI that can provide comprehensive information necessary for evaluation and can help with decision making. Through the explication and suggestion of information on the mechanism of toxic effects to safety assessment scientists, a subsidiary partnership system for risk assessment is ultimately to be a powerful tool that can indicate project-vector with data weight for the corresponding counterparts. To bridge the gaps between big data and knowledge, multi-dimensional thinking based on philosophical ontology theory is necessary for handling heterogeneous data for integration. In this review, we will explain the current state and future perspective of computational toxicology related to drug safety assessment from the viewpoint of ontology thinking.
In terms of the data for the safety assessment of pharmaceuticals, initial data management means how to accurately gather toxicological data, appropriately analyze their statistics, present it in summary tables and diagrams, and utilize for safety assessment with supporting systems for experts properly. Usually, the data are to be derived from regulatory safety studies (single / repeated administration toxicity, gene toxicity, safety pharmacology, reproductive development toxicity, carcinogenicity, etc.) under the application of related guidelines.
Computerizing systems have been initially developed under the application of Good Laboratory Practice (GLP) that can assure the integrity and quality of the obtained data with study management and data gathering / retrieval / reporting.
Subsequently, from the point of attrition during the development of drugs, safety evaluation in the early stage of drug discovery is essentially required as High Throughput (HTP) / investigative toxicity test in addition to the guideline following the type of regulatory toxicity test. These approaches have been applied for the selection of lead compound and candidate compound for clinical trial. On the selection of proper compound as lead compound, a computational system including Quantitative Structure-Activity Relationship (QSAR) is required by using archived database.
Furthermore, in the “Go” or “No-Go” decision of drug development, supportive utilization of computer system has come to be drawing attention as predictive analysis and mechanism analysis of toxicities, and risk assessment with utilization of comprehensive database.
Having practical risk assessment / management on predictive human safety evaluation as described above, it is important to construct a scientifically interpretable computational toxicology system. A pharmaceutical safety evaluator as a toxicologist who is facing practical decision does not need a data-driven Artificial Intelligence (AI) system that calls for the final consequence, but rather seeks the provision of comprehensive information necessary for evaluation and help with decision making. Furthermore, through the presentation and suggestion of information on the mechanism of toxic effects to safety assessment scientists, it ultimately builds a subsidiary partnership system that can result in extrapolating risk assessment and management. Finally, the system is required to provide the indication of project-vector with data weight / depth for the corresponding counterparts. Taking some notice that there are large, deep gaps between the big data and the knowledge for constructing a scientifically interpretable system, it is necessary to grasp a wide variety of computational toxicology systems as a matrix world and to develop multi-dimensional thinking based on philosophical ontology theory in order to handle heterogeneous data for integration. Ontology deployment has a machine-like informatics-oriented ontology field and a scientific thinking (philosophy-oriented) ontology field. In other words, bioinformatics of informatician would develop “informatics-oriented ontologies” based on Information Technology (IT) and scientists and ontologists would construct “philosophy-oriented ontology” based on scientific thought.
In this report, the current state and future prospects of computational toxicology related to drug safety assessment are addressed from the perspective of ontology thinking.
Transition of toxicological data gathering, management, reporting and safety evaluation with corresponding computer systems in the field of computational toxicology is shown in Fig. 1.
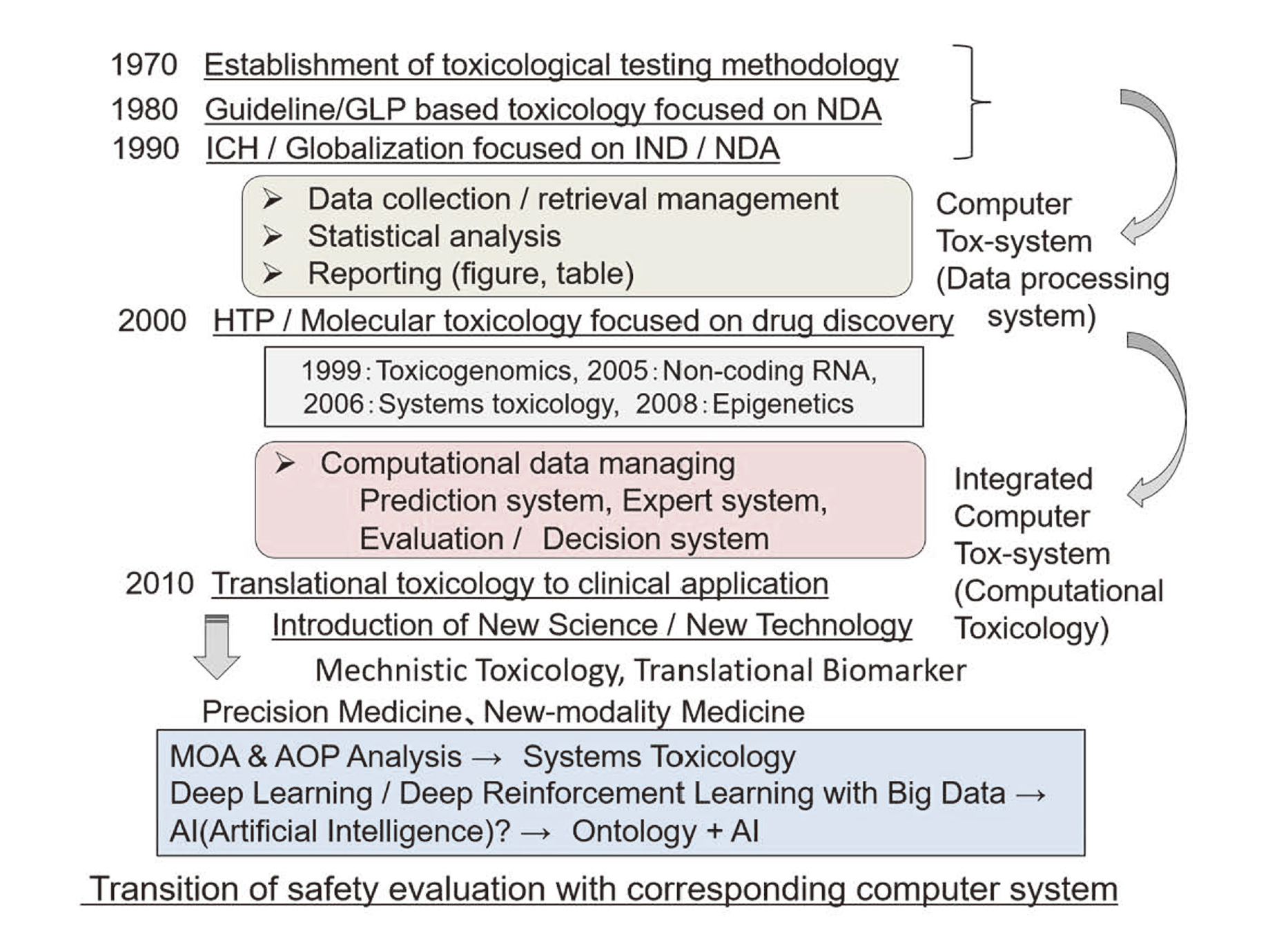
Transition of drug safety evaluation with corresponding computer system.
Implementation of drug safety study initially started as a regulatory science under the guidelines established in the legal regulation for New Drug Application (NDA) in the 1980s, focusing on the conduct of toxicological studies under the GLP regulation to guarantee the quality and integrity of gathered safety data (Horii, 1994). The enforcement of computer system at the time was mostly focused on accurate data collection, significant statistical analysis and proper reporting as tables and figures.
Thereafter, in the 1990s, these regulations and guidelines were globalized under the International Conference on Harmonization (ICH) of Technical Requirements for Registration of Pharmaceuticals for Human Use regulation, and safety assessment in nonclinical study at clinical application to humans (IND: Investigational New Drug) has been also pointed to consider in addition to NDA preparation. In parallel with these movements, investigative toxicological study with HTP evaluation in the early stages of drug discovery has started being introduced. Beginning in the 2000s, molecular toxicological approach has risen secondarily from pharmacogenomics drug discovery according to the progress of molecular biology, and new sciences such as toxicogenomics, systems toxicology, non-coding RNA regulation, epigenetics, etc. have been proposed as a matter of challenge. Based on the trends of these new sciences in the 2010s, translational toxicology from the search for biomarkers has been presented, and further innovative object of drug discovery has been begun to pursue a new modality of medicines such as precision medicine, regenerative medicine, gene therapy, etc.
The database concurrent with the progress of these new / innovative science and technology and its usability has been leading to the new type of computerized systems for safety and risk evaluation / management. These movements would mean to evoke the next generation of computational toxicology, namely AI in the field of toxicology.
System matrix of toxicological database and related computational system for safety evaluation on drug discovery / developmentCurrently used toxicological database and computational systems related to safety evaluation and risk management based on various safety assessment systems are shown in Fig. 2. From the viewpoint of computational toxicology to give a significant contribution for the safety evaluation of pharmaceuticals, initial clarification about the entrance/exit of total system configuration would be desired as following points (data-source, data & analysis, outcome): type of the source of data derived from investigative research study, non-clinical and clinical study, post-marketing survey study, and related literatures, scientific characteristics definition for data analysis and systematization for mechanistic safety evaluation in each computational database analysis, and usable outcome such as Mode Of Action (MOA) analysis, toxicological target setting, biomarker setting, selection of lead / candidate compound selection, go / no-go decision making, risk evaluation / management, etc (Horii, 2016).
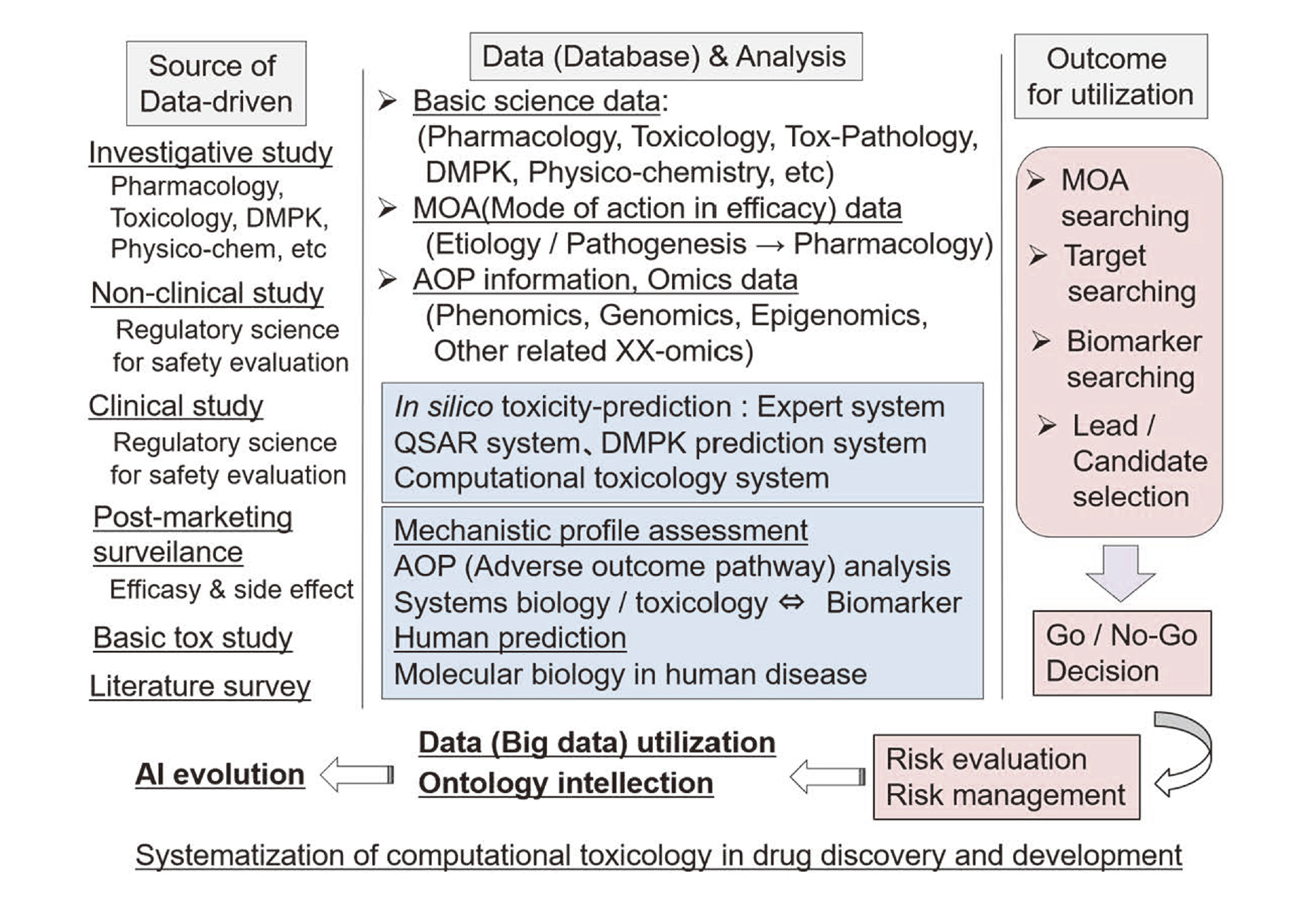
Systematization of computational toxicology in drug discovery and development.
Sources of utilizing data are to be derived from investigative study (pharmacology, toxicology, pharmacokinetics, physicochemistry), regulatory nonclinical study, regulatory clinical study, commercial survey (drug efficacy, side effects), literature survey, etc.
Data analysis is to be charged on comprehensive computational systems such as in silico prediction system, QSAR system, expert system, related computational toxicology system in specified science field, etc.
For human prediction, outcomes from data evaluation and toxicological mechanistic analysis should be referred based on Adverse Outcome Pathway (AOP) / MOA analysis, translational biomarker and systems biology / toxicology.
In any case, in order to achieve the ultimate goal of system utilization, it is necessary to clarify and define the entrance (data providing place) and exit (data utilization) for such matrix-like big-data and take it for the “go” or “no-go” decision during drug development, safety/risk evaluation/ management, mechanism elucidation, extrapolation to human, etc. In addition, we believe that ontology thinking would be indispensable for further development of AI approach in order to incorporate Human Intelligence (HI) as a philosophy-oriented ontology.
Depending on the user’s purpose, standing-point of system-matrix and weighing of each computational system would be changed in each demanding item of retrospective analysis, prospective analysis, mechanistic analysis, biomarker searching, challenge for coming new science and technology, or challenge for innovative medicine as a new modality.
Databases for safety assessment on adverse drug reaction are diverse, and defined as pharmacological form & predicting Adverse Drug Reaction (ADR) from pharmacogenomics / toxicogenomics (Tan et al., 2016), search by pathway analysis based on gene expression (Davis et al., 2013; Duan et al., 2014; Hardt et al., 2016; Hendrickx et al., 2014,), database search specialized for organ specific toxicity (Chen et al., 2013b; Teufel, 2015) (Table 1).

Taking the background of these data sources into consideration for safety evaluation, the following systems have been developed aiming at constructing QSAR model and computational prediction model: a search focusing on corresponding organs such as liver toxicity and QT prolongation (Chen et al., 2013a; Liu et al., 2015; Mulliner et al., 2016; Valerio et al., 2013), the predicting model (Wittwehr et al., 2017) mainly by regulatory toxicology of AOP -base, the prediction system by Deep Learning (Capuzzi et al., 2016; Mayr et al., 2016; Xu et al., 2015).
In any case, in these approaches based on the system construction, it was not possible to provide effective evidence directly linked to decision making at each stage from the early discovery phase of drug discovery to approval application as a total system.
Point to consider for the consolidation of significant safety evaluation and risk assessment / managementConsidering the usefulness of computational system consolidation, basic issues about the problems in the current existing system should be discussed in making a significant safety evaluation and risk assessment / management.
To set up a computer system primarily from the aspect of positioning and systematization of database on drug discovery / development, it is necessary to clarify what the system needs. Under the process of systematization, it is fundamental to understand the database itself and analyze the functions in the various systems as a system-matrix.
The correspondence analysis items to be addressed are as follows.
(1) Retrospective analysis, (2) Prospective analysis, (3) Mechanistic analysis, (4) Biomarker searching, (5) Challenge for new and upcoming science and technology, (6) Challenge for innovative medicine as a new modality
In the verification of safety, the timing of data provision as a trigger of toxic effect appearance and the availability of each system in view of the differences between the search purpose and its demand.
Systematization-needs for data-management is developed in each stage of drug discovery and development shown below (Fig. 3).
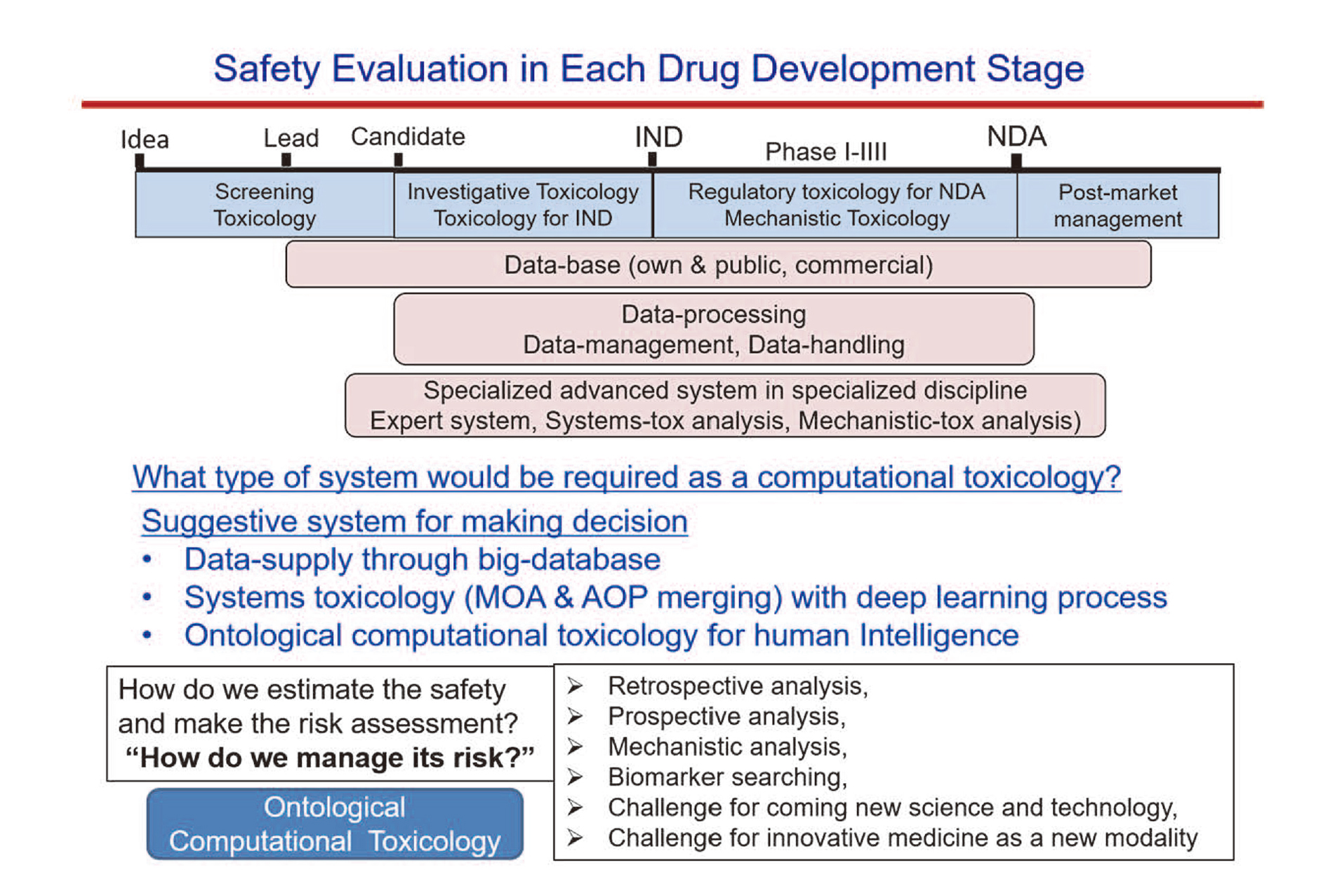
Related computational system in each drug development stage.
» Early screening stage of discovery: Search for possible causes and mechanism of toxicity in the upstream of gene fluctuation (omics data and machine learning), prediction by expert system, System construction for prioritization / ranking / optimization in early drug discovery
» Preclinical and non-clinical stage: Extrapolation study for mechanistic toxicology, Utility for go or no-go decision in drug development stage, AOP integration, Application for systems toxicology with pathway analysis
» Clinical phase with diagnostic analysis: Side effect information, Clinical data, Retrieval of related safety data (toxicology / pathology etc), Side effect database utilization
» Approval and preparation and application: Response to approval application and subject of evaluation by authorities, Information of post-marketing surveillance.
In provision of safety information and support for decision making, objective evaluation by computational toxicology based on a large amount of information would be indispensable. Based on the current computational toxicology (as one of AI tools) analysis, can we make a decision for safety / risk evaluation automatically? In reality, it is hard to simply say “yes, we can” because of the following unclear difficulties. The explanation for system-verification to meet the issues shown above is insufficient, although it can be said by an informatician that AOP and system toxicology are easy to understand by data visualization and presentation. In most cases, interpretation for safety / risk assessment and management is currently performed by related qualified scientists. That means that making a decision requires the knowledge of experts in various scientific fields. Practically, decisions are made by sharing knowledge and forming a consensus by a consolidated scientific group, and it also depends on the applicability to risk evaluation / management.
From the viewpoint of the difficulty mentioned above, AI development based on ontology thinking as matrix function seems to be required more in the future. In other words, what do we ask for artificial intelligence, including related computer system, in drug safety assessment? It is important to know that the system does not result in what it can do as a realistic matter such as biomarker mining from bulk data in searching, data mining of indicators, risk prediction based on probability, statistical correlation analysis by artificial intelligence, hypothesis generation from mathematical processing, risk classification based on pattern recognition, and clustering. The functions and capabilities of individual computerized systems with the aim of analysis of mathematical processing-based evaluation, quantitative evaluation, efficiency of risk detection, speedup, formality uniformity, etc. are clearly presented as a single thought of computational toxicology. However, since it is a two-dimensional or planar understanding of data itself, it is hard to say that it is sufficient for understanding data weighting / depth, direction of judgment, etc.
In any case, science-based safety evaluation should be in the theory of “Toxic effects are basically responses to xenobiotic substances, and expressed as triggering or additional accelerating adverse effects toward abnormal condition. Toxic effects as biological adverse responses, are interpreted as protective responses of living body, and the adverse effects caused by drugs are also considered to be protective responses” (Horii, 2010). These mechanistic toxicological thoughts should be standing on multi-disciplinary sciences including new sciences, such as molecular toxicology, etc.
Ontology derives from ontos as “existence” and logos as “rational account” or “knowledge” in Greek (Arp et al., 2015). In information science, with the explosion of data, how to integrate heterogeneous data is a key issue, and its attention is focused on ontology for use in data integration sharing and further knowledge sharing. The definition of ontology as “an explicit specification of a conceptualization” by Gruber is famous in AI area (Gruber, 1993). As a branch of knowledge engineering, ontology is defined as “a theory of concepts used as building blocks of an information processing system” (Mizoguchi, 2005).
Ontology enables us to support the systematization of diverse knowledge interdisciplinary in science. In bioscience, there are many biomedical domain ontologies such as Gene Ontology (GO) (Ashburner et al., 2000), Foundational Model of Anatomy (FMA) (Rosse and Mejino, 2003), and so on. A biomedical community Open Biological and Biomedical Ontology (OBO) Foundry aims for collaborative development of interoperable ontologies with high quality improvement (Smith et al., 2007).
From another perspective, types of ontology can be divided into “informatics-oriented ontology” and “philosophy-oriented ontology”. Informatics-oriented ontology is almost the same as what is called terminology or vocabulary set, which sometimes has a hierarchy, and practically means as tags or indexes to access and process a large amount of data in information science. Thus, there is no deep consideration on the validity of the classification without strictness against the criteria of the term selection (Mizoguchi, 2005). Philosophy-oriented ontology respects philosophical thinking and cares about rigid meaning of each term and axioms. It requires the inheritance and strictness in the relation between the terms. Philosophy-based domain ontologies are like OBO Foundry’s ontologies, which show the validity of each term by referring to the upper ontology, such as Basic Formal Ontology (BFO) (Arp et al., 2015). These philosophy-oriented ontologies are generally made manually by ontologists and domain experts in cooperation.
In drug discovery, informatics-oriented ontology may be available as tags to retrieve large amounts of data and search risks. However, there is an issue for safety assessment. In each case of evaluation, the meaning of each term should be made clear and scientific justification is needed.
As shown in Fig. 2, computational toxicology contributes to the safety evaluation in a way of providing useful data. One of the issues, however, is the so-called “silo problem” in which individual information is isolated for each database (DB). Most of the systems and DBs in computational toxicology have developed independently in their own specialties, which causes knowledge fragmentation. As a result, it is difficult for knowledge sharing or reusing.
Concerning safety evaluation, it is required for an integrated system. However, even if data acquired from various DBs are merely merged and stored together in one database, it is practically meaningless. Such data are so inconsistent that computers cannot process them adequately. Therefore, an intelligent integrated system is required to satisfy both capturing the whole picture from the early stage to the clinical stage and providing flexible information. As an advantage of ontology, ontology makes explicit the meaning and relationship between basic terms in common in the target world, which contributes to organizing the knowledge with consistency and supports knowledge sharing.
Ontology and its representation framework in drug safety assessmentWith respect to the drug safety assessment system, ontology can make explicit background knowledge, which has been implicit in each own expertise. Hence, ontology contributes to interdisciplinary developing knowledge infrastructure for systematizing the relationships between various kinds of data involved in drug safety. The ontological standpoint is that ontology can reuse the information in existing systems such as chemical structure databases etc., rather than “reinvent the wheel” (Arp et al., 2015). By providing metadata to existing data, it is possible to give meanings of data while retaining the original. However, concerning toxicity mechanisms, there was no consistent knowledge systematization. Yamagata et al. have focused on toxicological mechanisms for safety assessment and have been developing a toxic process ontology (TXPO) and its application system, TOXPILOT as toxic process interpretable knowledge system (Fig. 4 (Yamagata et al., 2019a, 2019b)).
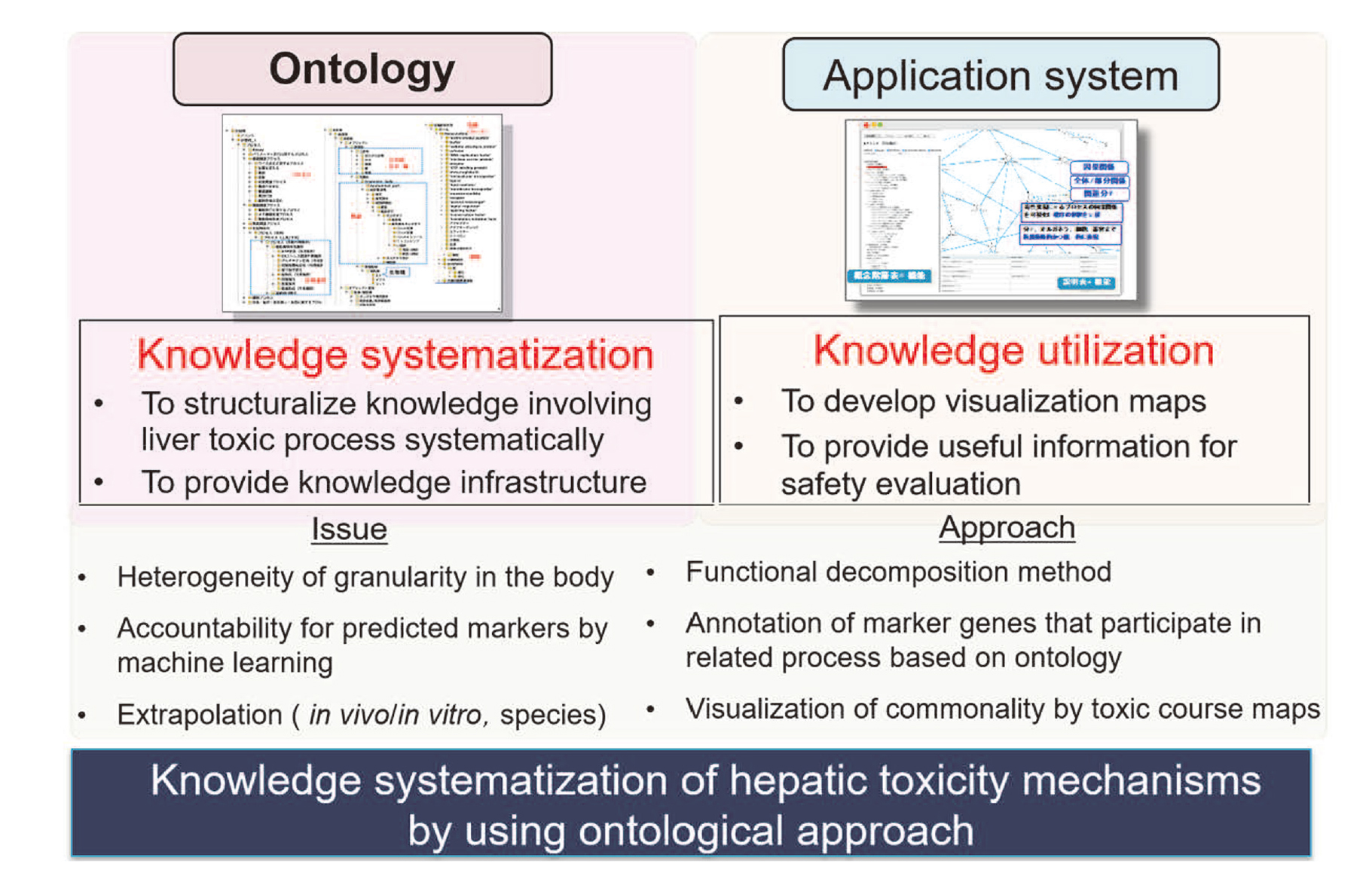
Knowledge systematization of hepatic toxicity mechanisms by using ontological approach.
Grasping the mechanism of toxicity is essential for the assessment of toxicity in drug development from drug discovery to clinical trials. However, toxicity mechanisms in organisms are diverse. Therefore, for building the knowledge systematically, the ontology has to cover terms from general to special appearing in toxicological science. Fig. 5 shows an outline of the toxic ontology. At the top, the concepts are classified into two categories: “Continuant” and “Occurrent”. The lower concepts of the “Continuant” are an organ, a cell, a molecule, an organism and the like. As a lower concept of “Occurrent”, “Process” is defined and is further classified into specific toxic processes and biological processes. A toxic mechanism can be grasped as a toxic course, i.e., a series of processes, which is the lower concept of the sequence of processes. Then, the ontology defines a dependent continuant that is dependent on some bearer such as role.
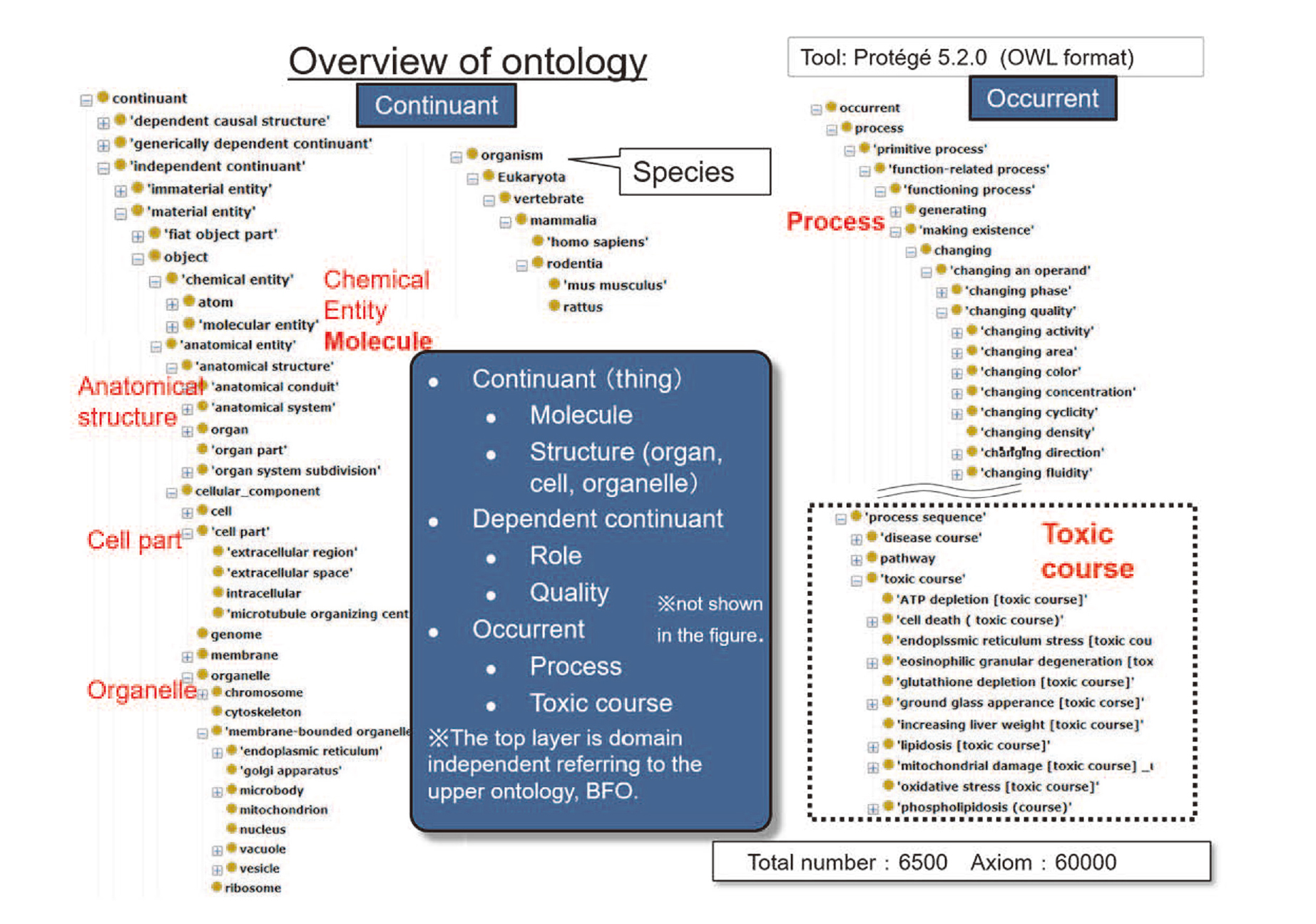
Overview of ontology.
As for the toxic course, it can be represented in terms of causal relationships of processes leading to the manifestation of the toxicity (Fig. 6.) To clarify the mechanism, it is essential to explicate how cells perform defense functions against the toxic action in the course of toxicity. By introducing a systemic decomposition approach in the engineering framework (Kitamura et al., 2004), a biological functioning process can be decomposed into several sub-processes of the system components. For example, in the course of endoplasmic reticulum (ER) stress, the Unfolded Protein Response (UPR) can be said to be “controlling the unfolded protein quality” as the cellular defense system. Furthermore, the UPR process can be decomposed into sub-processes of organelles as system components: “unfolded protein refolding” in the ER, “unfolded protein degradation” in the cytoplasm, and “translation attenuation” in the ribosome. Here, molecules can be represented as participants of the specific process. For example, Bip participates in the protein refolding process and has a role as “chaperone.” In this way, this framework can cover concepts from the molecule to the cell level across granularity, and consistent knowledge systematization contributes to support interpreting the toxicity mechanism.
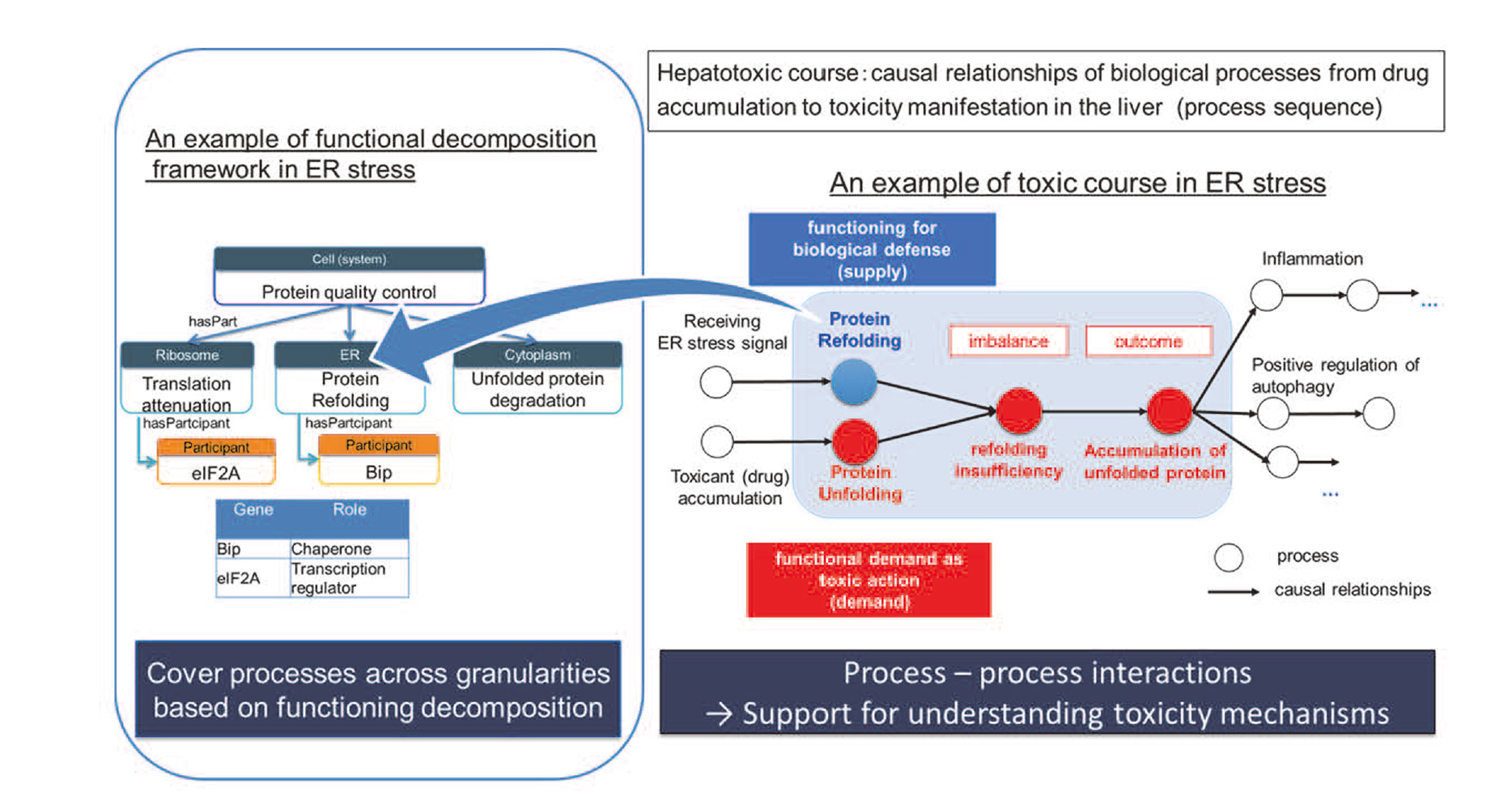
Unified description framework of toxic course and process decomposition.
As an application of ontology, development of knowledge infrastructure will support the integration of various heterogeneous data of existing DBs in computational toxicology.
Considering drug safety assessment in the stages of drug development (Fig. 3), key issues for the computational system are “what type of system would be required as computational toxicology?”, “how do we estimate the safety and make the risk assessment?”, and “how do we manage the risk?” As a suggestive system for making decisions during the course of drug development, the following points are to be addressed: data-supply through big-database, systems toxicology (MOA & AOP merging) with machine learning process, and ontological computational toxicology for human intelligence. Necessary analyses for data processing would be retrospective / prospective analysis, tox-mechanistic analysis, biomarker searching, and challenging for coming new sciences and new modality medicine.
This section introduces the case examples by using the ontology-based knowledge system.
Examples:
1. Toxicity mechanism search
2. Search from chemical structure
Possible Example:
3. Challenge of the inference of patient groups who have the possibility of idiosyncratic toxicity expressions
The ontology-based knowledge system is possible to illustrate how factors can be related in the body according to the user’s viewpoint, and this system also enables to explicate the course of the toxicity manifestation. This knowledge system is possible to give accountability to computers, which will support the right decision for safety management of risk reduction. Using the system, integrating various kinds of heterogeneous data from basic science to clinical medicine will also contribute to elucidate the idiosyncratic toxicity. Example 3 is a prospect and challenge for the inference of patient groups who have the possibility of idiosyncratic toxicity expressions.
1) Toxicity mechanism searchToxicity mechanism is essential in all drug development stages. Therefore, having the ability to represent general toxicity mechanisms is the first step in knowledge integration system development. In the course of drug excretion, metabolites often interact with biological components and lead to toxic manifestation.
Fig. 7 shows an example of the visualization of the acetaminophen metabolite, N-acetyl-p-benzoquinon imine (NAPQI), and related process in the knowledge system. In the course of cytochrome P450 (CYP)-mediated acetaminophen metabolism, the intermediate NAPQI is generated as electrophile species. Then, increased demand for glutathione conjugation leads to an imbalance between glutathione production and degradation, and as a result, glutathione depletion can occur.
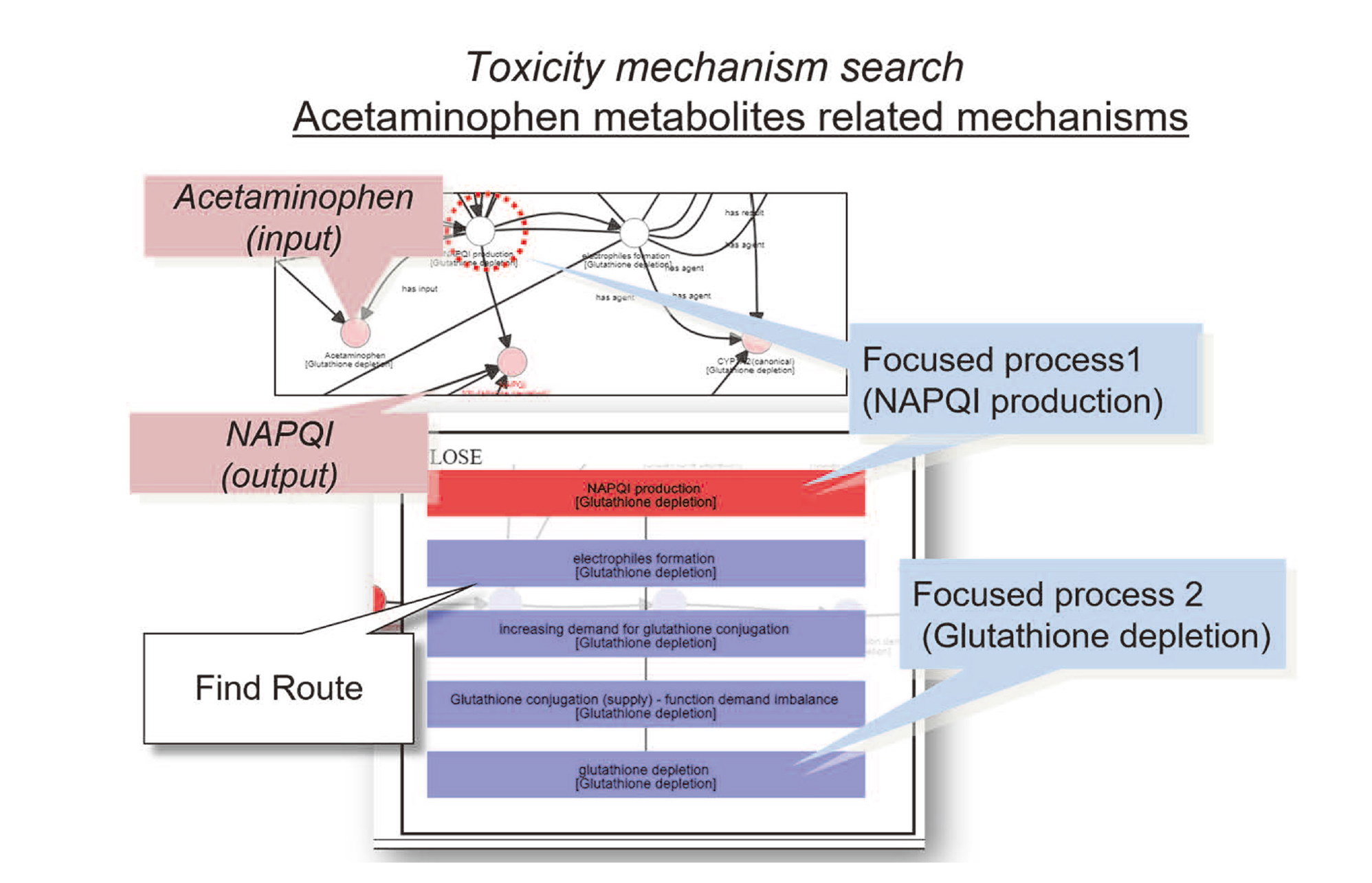
An example of toxicity mechanism search related to the acetaminophen metabolites.
As an advantage of ontological utilization, ontology can find commonalities among multiple toxic courses by generalization. Knowledge system can show that both the metabolites of acetaminophen and carbon tetrachloride affect common processes such as oxidative stress, Reactive oxygen species (ROS) production, and mitochondrial damage, which may lead to necrosis.
In this way, the system makes it possible to acquire more knowledge from the focused point by finding commonalities and differences accordingly and following the relationships between data derived from different DBs. In cooperation with omics data and machine learning, knowledge system also affords the acquired explanations on how predicted genes are involved in the specific toxic course such as ER stress and how each gene plays a role in the specific process (Fig. 8).
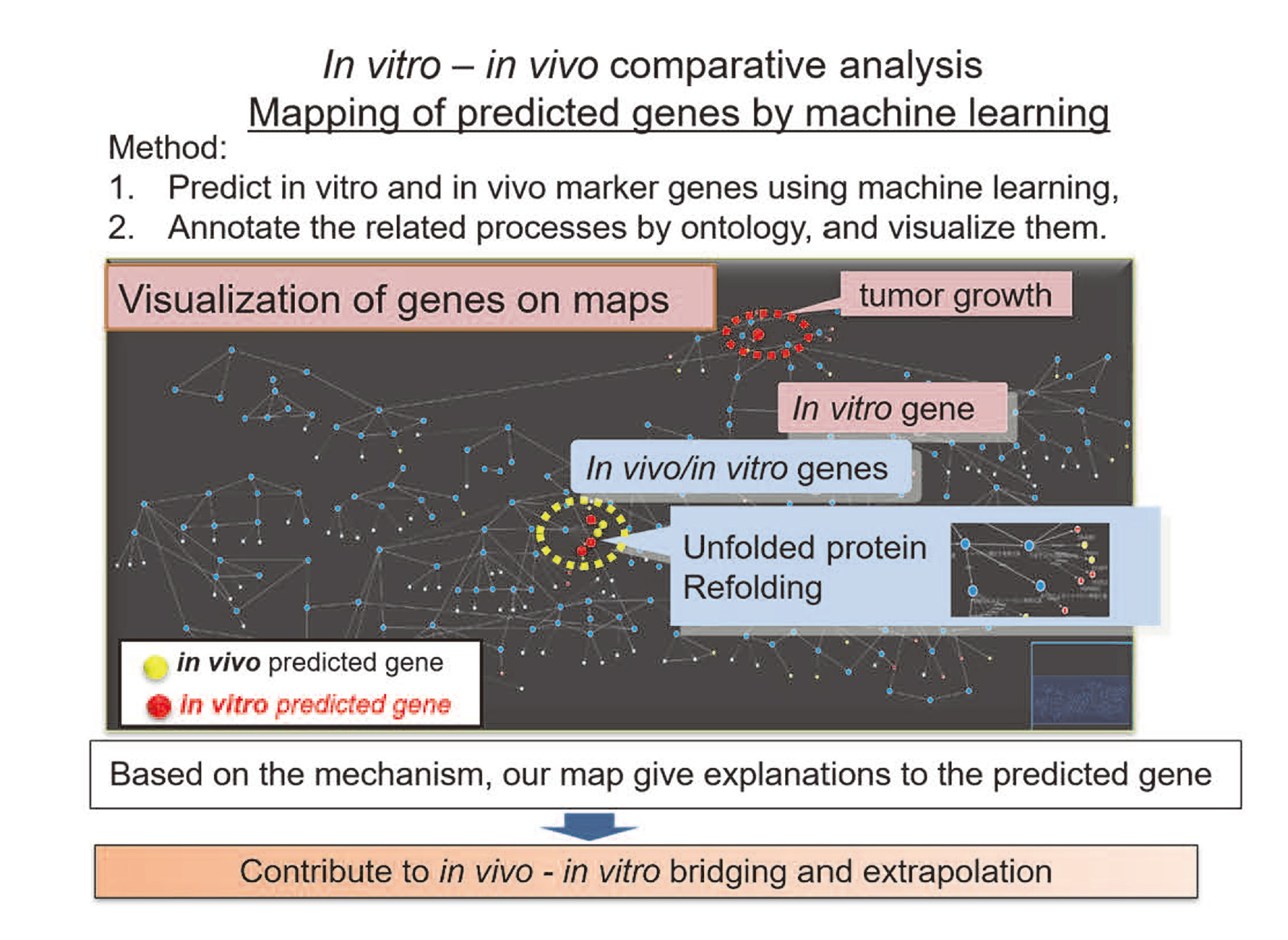
In vitro - in vivo comparative analysis: mapping of predicted genes by machine learning.
So far, concerning computer systems related to structure, various tools, for instance, QSARs and quantitative structure-toxicity relationships (QSTRs) models, have been developed for the prediction of correlations between structure and activity/toxicity. However, at present, concerning toxicity mechanisms, there is a large gap between physicochemical properties and biological activity. These tools do not answer why correlation occurs and how processes occur during the toxicity manifestation.
Ontology-based knowledge infrastructure will overcome this problem and give accountabilities between them according to the toxicity mechanisms.
For example, using knowledge system, when Cationic Amphiphilic Drugs (CADs) are searched, the following related processes are represented:
• Input: CAD,
• MOA: phospholipase inhibition by binding to lysosomal membranes,
• Related toxic course: Phospholipidosis
• Core process in toxic course: lysosomal phospholipid accumulation in phospholipidosis
• Processes in phospholipidosis
• Occurs in organelle: lysosomal degradation, lysosomal enzyme release
• Occurs in cell: necrosis, increasing apoptosis
• Occurs in tissue or organ: inflammation, liver dysfunction, tumorigenesis
• Pathological findings: Lysosomal myelin-like structure formation [hepatic electron microscopic finding], macrophage-derived foam cell increase (toxicity (severe))
Thus, the ontology-based system makes it possible to interpret the mechanism comprehensively from chemical structures, biological function, toxic process, or the findings. In the future, in terms of toxicity screening, for example, by combining information about chemical structure, log P (lipid solubility), gene expression, and toxic course path similarity, it is possible to make structural reclassification based on kinds of biological processes.
As another example, with regard to the path where toxicity expression is strong, by searching for alternative pathways that weaken toxicity, researchers can newly investigate chemical structures that target molecules expressed in the upstream processes of the alternative pathways. In this way, the system will be useful for comprehensive analysis for risk reduction.
3) Challenges to inference of patient groups who have a possibility of idiosyncratic toxicity expressionsResponses to drugs are not often unimodal among individuals. One of the causes is based on genetic mutations such as Single-Nucleotide Polymorphisms (SNPs). Due to the involvement of various factors in the development of toxicity, no straight way exists to identify patients with drug idiosyncrasy. If appropriate patient groups with the possibility of specific toxic expressions can be selected, it can be applied to technologies such as the establishment of iPS cells and can be expected to progress to safety prediction and management. Ontology could support data annotation by making metadata to existing data, providing control vocabulary, and explicating the relationship between data. By integrating and analyzing across various background data of patients such as genome data, clinical test, complications, medication history, race, gender, and the like, ontology is expected to find commonalities and differences in the latent factors and make them explicit before the onset of toxicity on patient groups.
The following is an example of how a computer can infer the possible susceptible population who may have toxicity expression similar to the liver damage caused by troglitazone:
Firstly, from genomics data, concerning troglitazone, patients with GSTM1 and GSTT1 deficiency are selected. Next, from Chemical Entities of Biological Interest (ChEBI) ontology (Hastings et al., 2013), using information that troglitazone (CHEBI:9753) is categorized into thiazolidinediones (CHEBI:50990). Then, the system searches drugs (e.g., pioglitazone (CHEBI:8228) and rosiglitazone (CHEBI:50122)) classified to the thiazolidinediones. After that, the system queries the electronic health record DB, which stores the patient’s background information and selects the patients who take thiazolidinedione drugs. Finally, by using all linked data, the system extracts patient groups, who are both taking thiazolidinediones and having SNPs of GSTM1 and GSTT1, as candidates for drug-induced idiopathic injury.
Concerning GSTM1 and GSTT1, the system extracts related information from the toxic course that these genes are involved in glutathione depletion and drugs such as acetaminophen are included in the course of glutathione depletion. Then, in hospitals, when acetaminophen is prescribed to the patient groups who have chronic-use of thiazolidinediones, the integrated system alerts that glutathione depletion is enhanced and the possibility of drug-induced toxicity like troglitazone exists. Thus, the system will contribute to the inference of the toxicity derived from the interactions with concomitant drugs.
In a limited area of safety assessment such as QSAR analysis, target organ safety estimation etc., each computational toxicology system can lead some suggestive answer to the aimed issues separately. Their corresponding items for the assessment / evaluation are definition of toxicological target organ / tissue /cell, severity of adverse effect, reversibility, indication of adversity, No Observable Adverse Effect Level (NOAEL), Lowest Observed Adverse Effect Level (LOAEL), etc. These can make a contribution to set lead compounds up and also suggest an appropriate candidate compound for clinical application, which data were based on investigative toxicological data derived from many exploratory toxicological studies. In the early stages of drug discovery, a setting of the toxicological target other than that of the anticipated drug efficacy is challenging. The toxicological target site varies depending on the type of drug, and the granularity level of the target is also different. For example, some may affect mitochondria, while others may stress the endoplasmic reticulum. Moreover, as an example of a larger level, fibrosis may occur in the liver tissues. In order to define the target structure appropriately, it is necessary to capture the biological phenomena across various granularity from an organelle, cell, tissue, and to an organ. Ontology makes explicit the whole picture of the target world across granularity. By systematizing and formalizing the part-of (whole-part) relationship of a structural/functional abnormality with consistency, we can examine how cellular damage can influence tissues and organs, why the damaged cell affects the organelle’s function, and so on.
As another contribution, ontology can provide a suggestion of mechanistic toxicology by specifying processes and causal relationships between them based on careful consideration. Ontology also contributes to reversible and irreversible judgment since we can distinguish intrinsic cause and a derived result (e.g., parameter change in the blood flow reduction with occlusion) by tracking causal relationships between processes. In addition, by prospective analysis of the causal relationship between processes, it is useful to presume the side effects and its severity, which supports safety management and planning to the prevention of severity.
Here, regarding the evaluation of NOAEL, the background data accumulated for each facility is sometimes different, which leads to lacking unified data. In such a case, ontology can play a role as data schema and can enhance data standardization, allow data compatibility, and contribute to quality assurance.
At the timing of safety evaluation for submission and approval, the addressing of human prediction / relevance and risk assessment / management in patients is indispensable. To proceed with this issue, we should address “what kinds/types of data should be referred for the estimation of human risk assessment and management?” In order to increase extrapolation, bridging the gap between clinical data and basic research data is essential. The key idea is that ontology is domain independent and takes a neutral position. For human prediction, reference data in each species is indispensable for comparative analysis, and their relation in ontology hierarchy suggests both the inherent nature of each species and commonality sharable across species. By systematizing knowledge, ontology gives opportunities for consideration as to whether the similarity is derived from a structure or functional aspects, or how commonality or difference is born. Machine learning approach to finding biomarkers is also attractive for toxicity prediction. Ontology enables the improvement of accountability for biomarkers based on the participating process or molecule role according to the relevant concepts. In risk management, planning corresponding to the characteristics of each patient population is desirable. Depending on the levels of biological defense, the toxicity severity expressed in patients varies. However, there are multiple defense systems from an organ/ cell, to a molecular level in the body. To avoid misunderstanding risks, we should discuss multidimensionally and comprehensively from a diverse scientific viewpoint such as pharmacokinetics, pathology, and molecular biology. Ontology highlights the background knowledge of experts and turns implicitness into transparency by exchanging and sharing knowledge.
For corresponding to new modality derived from the approach of new sciences, we should address “what levels of new sciences should be picked up for further analysis?” In order to discover new knowledge, linking multidisciplinary knowledge is crucial. For instance, toxicogenomics was born as a new discipline in which toxicology unites with genomics, and its technology is continually changing. Therefore, in order to appropriately associate a wide variety of technologies, knowledge accumulation is important. However, it is a hard task to interpret each expert’s knowledge and convert it into a reusable form. An ontological approach enables to explicate the meaning of data and systematize the relevant information, which contributes to supporting knowledge interpretation across multiple domain experts and developing knowledge infrastructure. If knowledge sharing is accelerated, it may lead to creating new biomarkers, and its toxicity prediction marker may be applicable to diagnostic markers of rare diseases, and vice versa.
Every compound has the potential to induce adverse effects. With changing the scope freely and making knowledge interoperable among experts, the ontological approach would make a new challenge to provide a good indication for a go or no-go decision during the several stages of drug development. In order to realize the scenario shown in the previous section, it is necessary to systematize the knowledge consistently with the explicitness of each viewpoint. Knowledge integration is more than just connecting a large amount of data. For integrating various kinds of knowledge and performing higher-order knowledge processing, it is crucial to systematize knowledge based on deep consideration, where knowledge can be extracted in an appropriate form to be shared. By taking the ontological approach as a core technology and its application system of a knowledge base linking the data to various other ontological aspects and databases such as structure DBs and clinical DBs, it will contribute to the progress of computational toxicology (Fig. 9).
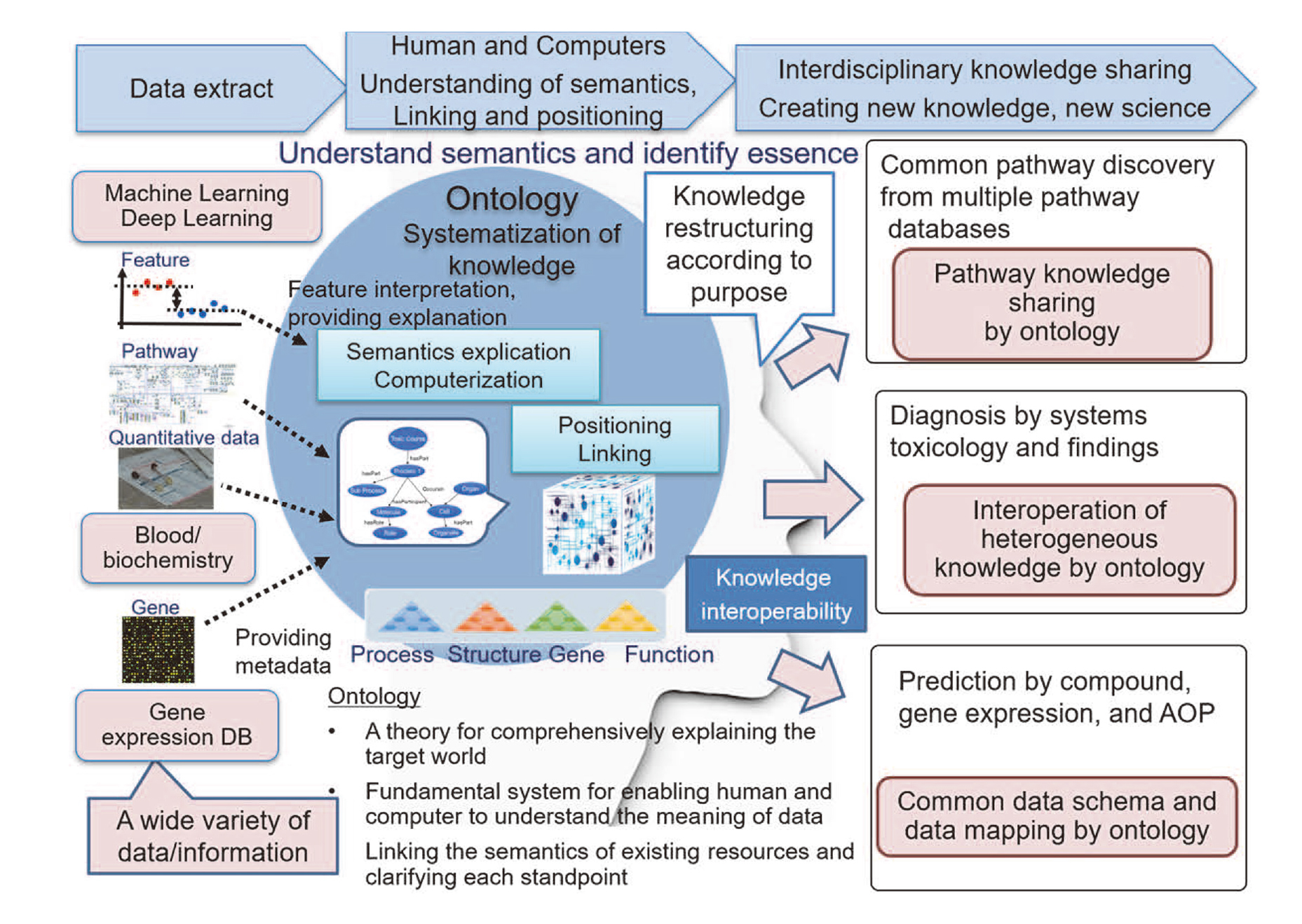
Ontology-based toxicity data integration.
Pursuing only practical and efficient big data processing by AI is not all there is in computational toxicology. Behind the expectations to AI that computers can process all data automatically, there might be a tranquil escape from facing up to the responsibilities for safety assessment/evaluation. To extract information appropriately from big data and to convert it to scientifically interpretable knowledge for safety assessment, deep thinking is essential from a comprehensive variety of perspectives on how to face safety issues scientifically to decrease risks for humans. That is intelligence, i.e., the ability to know its essence.
Ontology stands at the unique position bridging between human and computers. Rather than the dichotomy of AI and HI, ontology incorporates both and enables communication with each other (Fig. 10). For artificial intelligence, to exhibit intelligence, ontology supports analysis and interpretation comprehensively and navigates through appropriate knowledge flexibly with consistency. According to the purpose of each toxicological study, ontology can change weighting and provide accountability to the data. For human prediction, it is possible to accumulate knowledge in computers systematically and share knowledge across disciplines. Therefore, ontology assumes a crucial role to build a fundamental knowledge infrastructure of computational toxicology. Ontology enables the flexible conversion between data, information, and knowledge. Taking both advantages of computers and humans, knowledge interoperability between them will contribute to the progress of computational toxicology.
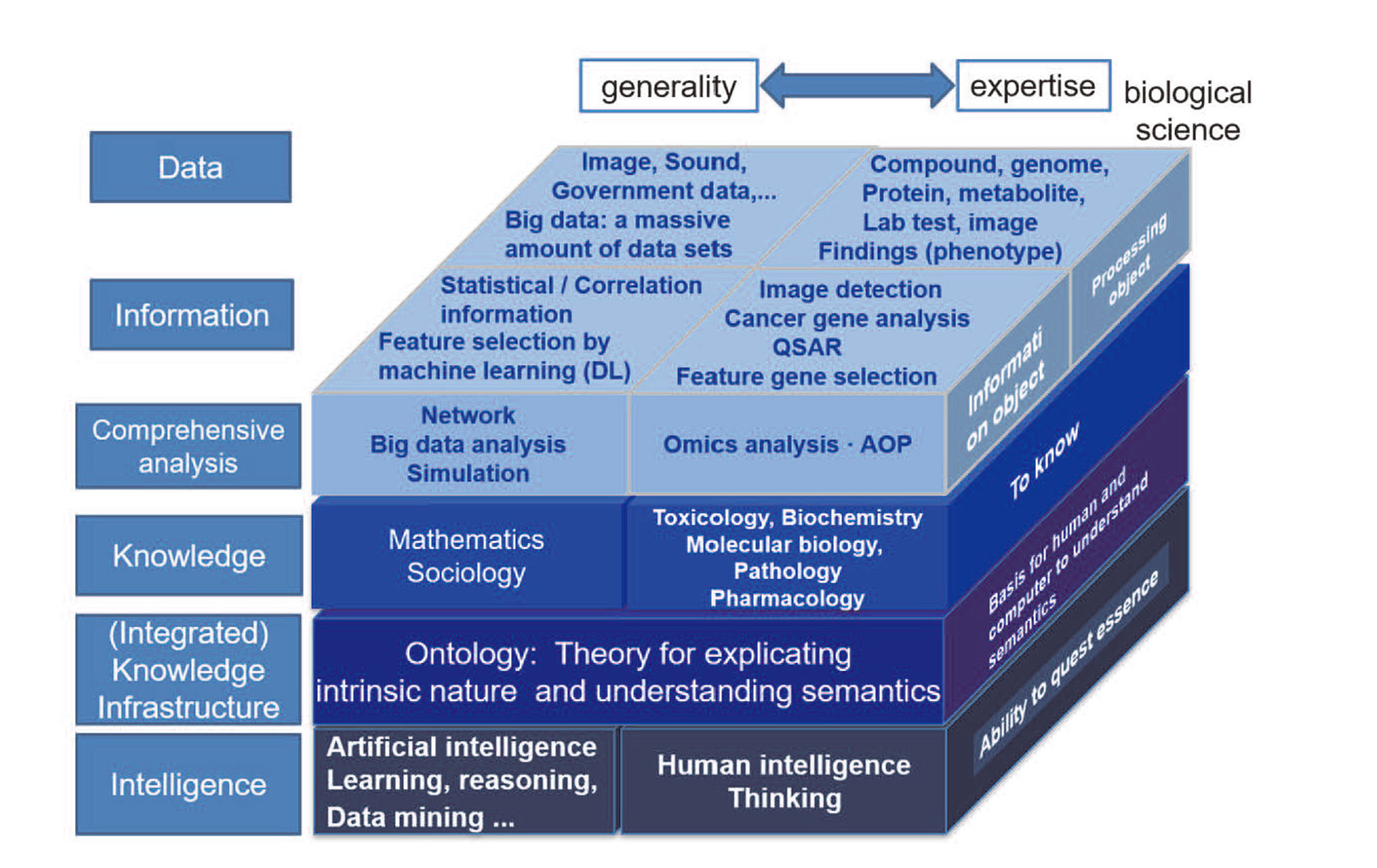
Role of ontology in Data, information, knowledge, and intelligence layer.
The authors would like to express their deep thanks and appreciation to Emeritus Professor Yasushi Yamazoe (Tohoku University) and Professor Toshiyuki Kaji (Tokyo University of Science) who kindly spared the time to review this article.
Conflict of interestThe authors declare that there is no conflict of interest.