2017 Volume 6 Issue 3 Pages S0072
2017 Volume 6 Issue 3 Pages S0072
Although matrix-assisted laser desorption/ionization (MALDI) mass spectrometry is one of the most widely used soft ionization methods for biomolecules, the lack of detailed understanding of ionization mechanisms restricts its application in the analysis of carbohydrates. Structural identification of carbohydrates achieved by MALDI mass spectrometry helps us to gain insights into biological functions and pathogenesis of disease. In this review, we highlight mechanistic details of MALDI, including both ionization and desorption. Strategies to improve the ion yield of carbohydrates are also reviewed. Furthermore, commonly used fragmentation methods to identify the structure are discussed.
Matrix-assisted laser desorption/ionization (MALDI) is one of the most important soft ionization methods for biological mass spectrometry.1,2) Several unique aspects of MALDI, such as the production of primarily singly-charged species and the high tolerance against contaminants, make MALDI a promising tool to explore unknown structures in a complex system.3,4) It has become one of the most well-established approaches in proteomics owing to its high sensitivity, low sample consumption, and minimal sample handling. With the growing interest in biological roles of carbohydrates, the structural identification of carbohydrates has also been attempted by MALDI mass spectrometry. However, compared to proteins, carbohydrates have lower ionization efficiency in MALDI, making carbohydrate analysis difficult. In fact, the lack of knowledge about the reaction mechanism in MALDI has restricted its applicability in analytical chemistry.
Carbohydrates (also called glycans) play important roles in mediating intercellular and intracellular functions such as protein folding, signal transduction, and cell–cell interaction.5–7) In particular, glycans become attached to proteins during glycosylation, which is one of the most ubiquitous protein post-translational modification (PTM). Aberrant glycosylation has been found to indicate the development and progression of cancer.8) Therefore, expression profiles of glycans are utilized as biomarkers for diagnosis and prognosis of cancers as well as therapeutic evaluation.9–13)
The “holy grail” of glycoanalysis is to be able to characterize the oligosaccharide heterogeneity at each glycosylation site on a protein or proteoglycan.14) The structural characterization of biologically significant glycoconjugates also presents opportunities for the synthesis of specific glycans to provide new therapeutics, such as carbohydrate-based vaccines for cancer.15) However, owing to their highly branched structures, thermally labile properties, and lower ionization efficiency, improvement of ion abundance as well as the structural determination of carbohydrate identification and sequencing with MS are still challenging. The study of ionization mechanisms is therefore crucial to the facilitation of efficient analysis of carbohydrates.
In this review, we focus first on the mechanisms of MALDI, including ionization and desorption processes. The discussion of underlying ionization processes of carbohydrates and methods for improving structural analysis of carbohydrates are reviewed, with special emphasize in the analysis of glycans in glycopeptides and glycoproteins. For further advances in carbohydrate analysis, please also refer to the recent review articles and books.16–21)
Matrix-assisted laser desorption/ionization comprises photochemical (ionization and fragmentation) and mechanical (desorption) events. In MALDI, analytes are incorporated into organic matrices in a solid or viscous phase (ionic liquid matrices). A pulsed laser irradiates the sample to facilitate the ionization/desorption reactions. During laser excitation, matrices take up and redistribute the laser energy for desorption and ionization of analytes. In this section, we first review critical factors determining efficiencies of ionization and desorption. Models proposed for ionization and desorption processes from the perspectives of both experimental observation and theory are discussed. After the review of general MALDI mechanisms, the specific aspects of ionization mechanisms on carbohydrates and methods that improve ion abundances of carbohydrates are reviewed.
Ion production in MALDIOwing to the high complexity of the MALDI process, a single ionization model that could elucidate phenomenon universally to every MALDI experiment is highly unlikely. Ionization environment varies among different matrix systems as well as excitation conditions.22) For investigating mechanisms in MALDI, the ion-to-neutral ratio (ionization efficiency) is normally discussed.23–26) Typically, ionization efficiency differs across types of analytes. The ionization efficiency for molecules with higher proton affinity than matrices, such as peptides and proteins, is 10−3–10−4.27–32) For analytes with lower proton affinity, such as carbohydrates, the ionization efficiency drops by a few orders of magnitude to 10−7–10−8.23,33,34) A better understanding of mechanisms leads to improvements in the ionization efficiency and increases its effectiveness toward more fields. Over the last three decades, fundamental ionization mechanisms have been systematically studied by manipulating one or more parameters among different matrix systems.22,35) Effects of the incorporation of analytes into matrix solid,36–40) matrix-to-analyte ratio,41,42) solvent constituents,43–47) laser excitation conditions,22,48,49) and the crystal morphology of samples47,50–55) on ion production were investigated.
A prerequisite of successful MALDI processes is absorption of laser energy by matrices. The optimal MALDI-MS sensitivity is generally achieved when the utilized laser wavelength corresponds to the high optical absorption band of the solid-phase matrices.56) After photo-absorption, a two-step ionization process suggests that ionization of matrices is as an initial ionization step, followed by ion–molecule reactions to produce charged analyte species.35,57) The second step is less controversial, and ion yields of analytes can be estimated by relative thermodynamic properties between matrices and analytes.58–61) However, how matrices are initially ionized remains open to debate. Two major arguments of postulated initial ionizations include 1) photoionization in the gas phase (Eq. (1)),35,62–65) and 2) thermal energy promoted ionizations in the condensed phase (Eq. (2)).34,66–74)
 | (1) |
 | (2) |
Contrary to the two equations, the lucky survivor model suggests that matrices and analytes are pre-charged in solids.78,79) In this model, the laser energy overcomes ion-pair interactions to free analyte ions from their respective counter ions. It should be emphasized that a single ionization mechanism that is applied universally to elucidate all MALDI observations is highly unlikely owing to different photochemical properties of matrices as well as excitation conditions. Contributions of different ionization channels need to be considered even for a single matrix system in Fig. 1.22)
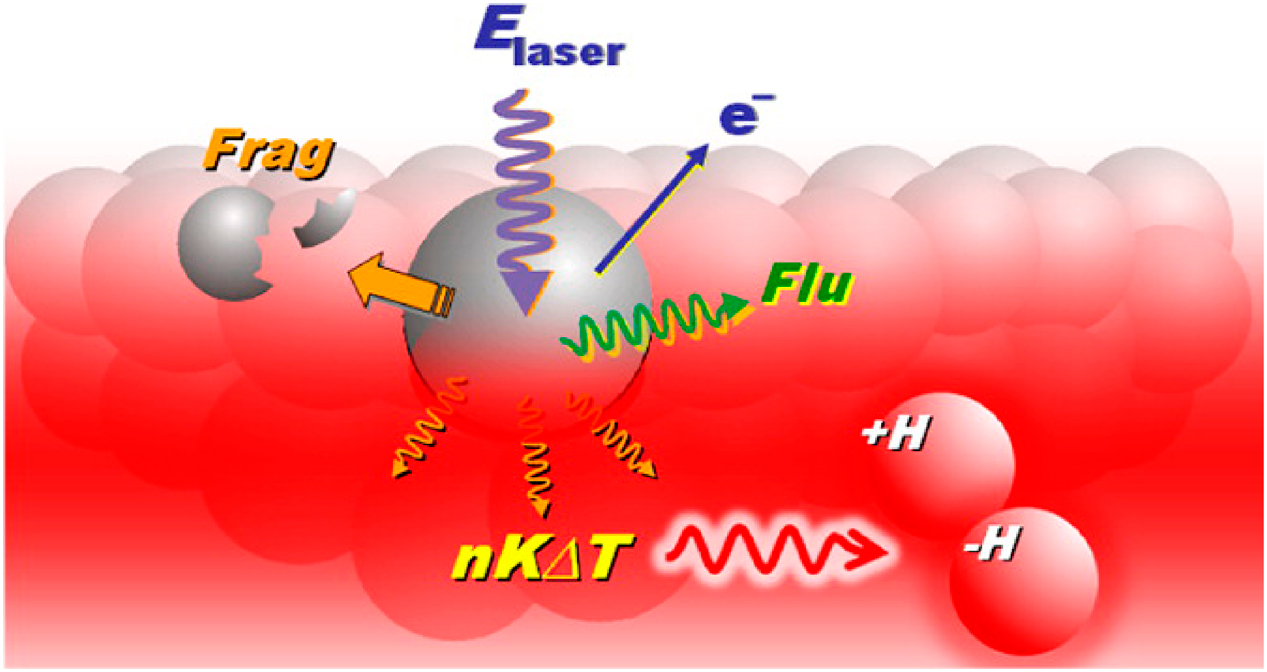
Desorption in MALDI is the phase transition of samples from the condensed phase to the gas phase. It is a prerequisite for molecules to be detected by MALDI mass spectrometry. Desorption processes, including thermal desorption occurring at low and moderate laser energy density to phase explosion happening at high laser energy density, were studied quantitatively on both experimental and theoretical bases.22,80) Results of these studies suggest that factors affecting the desorption efficiency can be classified into two categories: excitation parameters36,81–86) and intrinsic properties of samples.51,87–92) Generally, desorption efficiency increases when MALDI crystal size decreases51) and the system temperature increases.87–92)
On the basis of factors influencing desorption efficiency, mathematical equations as well as computational methods, such as molecular dynamics (MD) simulations, have been used to quantitatively describe the desorption process.86,93,94) As shown in Fig. 2, Zhigilei et al. utilized MD simulations to describe desorption of MALDI after laser irradiation.95) However, most of the aforementioned desorption models simplified the impact of ionization, which deviates from the fact that MALDI relies on the complex interplay of both desorption and ionization. By utilizing transition state theory to derive desorption rate, our group developed a comprehensive model that both considered desorption and ionization to predict ion abundance as a function of laser fluence,69)
 | (3) |
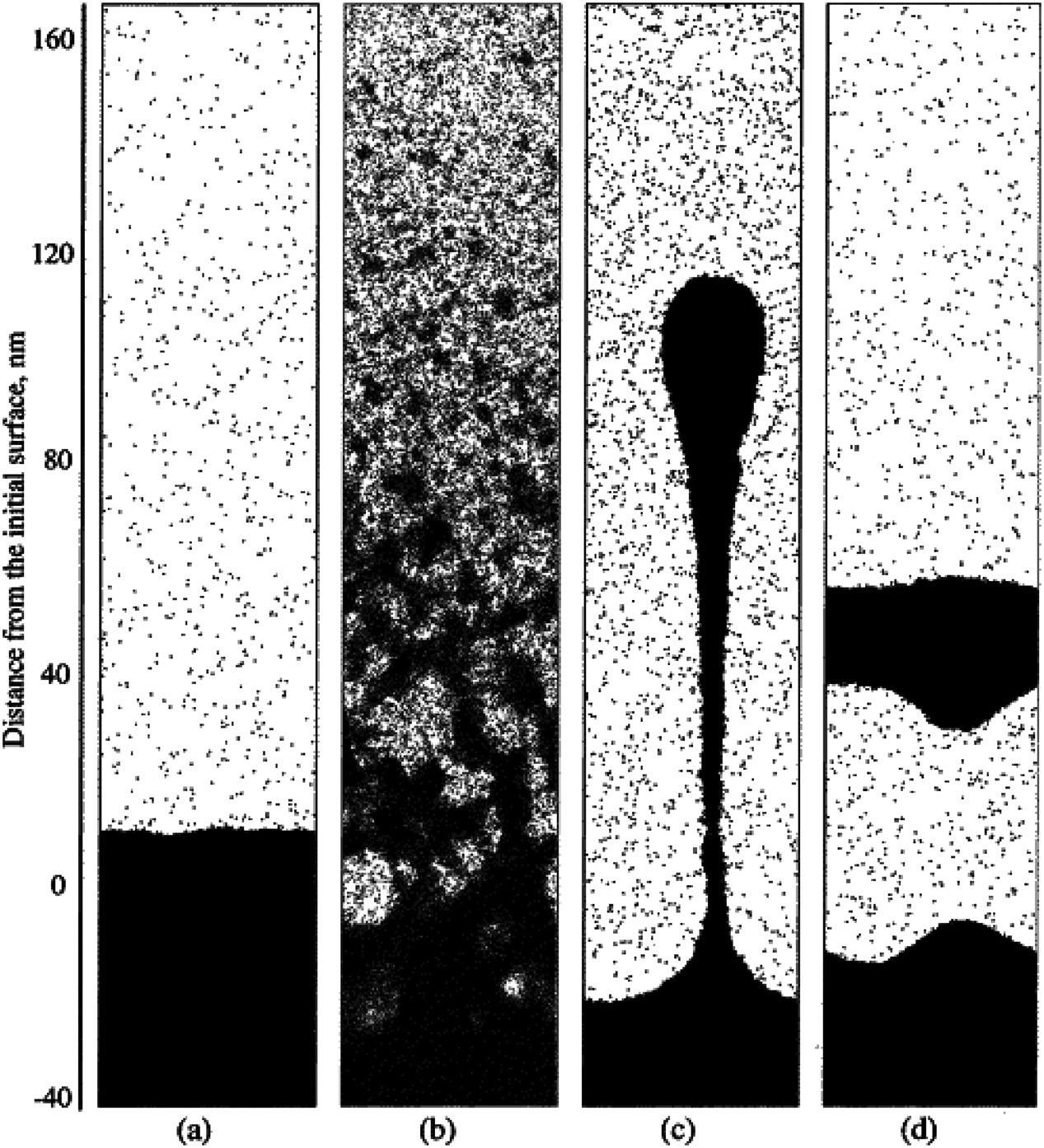
System temperature is a particularly important factor because it not only affects desorption efficiency but also ionization/fragmentation processes. The predicted system temperature varies with respect to experimental conditions, such as the type of matrices, energy densities, and irradiation wavelengths. Many efforts have been made to evaluate system temperatures of MALDI, such as predictions by theoretical models,62,69,70,86,96) MD simulations,32) the measurement of infrared emission of the system based on the theory of black body radiation,22,91) and the use of thermometer molecules as probes of internal energy.87,88,90,92) Although evaluations by different methods have certain inconsistencies, a consensus of transient MALDI temperature might reach ∼1000 K, which is detrimental to thermally labile carbohydrates.
Mechanistic studies of carbohydrate ionization in MALDIUnlike proteins or nucleic acids that are protonated or deprotonated, carbohydrates are predominately ionized by forming adducts with alkali metal ions (mainly sodium and potassium ions) in MALDI.97,98) Several groups have attempted both experimentally and theoretically to understand ionization mechanisms of carbohydrates in MALDI. Mohr et al. found that the binding affinities of proton and alkali metal ions to carbohydrates followed the order: H≪Li<Na<K<Cs.99) Since carbohydrates are incorporated into the matrix solid, the relative binding competition between carbohydrates and matrices needs to be considered. The binding affinity of protons to carbohydrates is lower than that to commonly used matrices such as 2,5-dihydroxybenzoic acid (DHB) and 2,4,6-trihydroxyacetophenol (THAP).30,100,101) In contrast to the proton affinity, the sodium affinity of carbohydrates is higher than that of matrices.98,101,102) Thus, it is energetically favorable for carbohydrates to form the adduct with alkali metal ions rather than with protons. A reaction model proposed by Lee et al. suggests that the increase in thermal energy can effectively induce the production of alkali ion adducts of carbohydrates.103) They suggested alkali metal ions were generated from the dissolution of salts at high-temperature MALDI condition. Those alkali metal ions attached to nearby matrices and analytes during desorption. However, mechanisms about how carbohydrates are alkalized are still unclear.104)
Ion yields of carbohydrates are related to multiple parameters, such as reaction enthalpies for forming adducts with alkali metal ions, volatility, and structural stabilities.101,105) Owing to the thermally labile nature of carbohydrates, Chen et al. reported that low ion yield might be the result of rates of fragmentation and/or desodiation (loss of sodium ions back to its neutral form) being faster than sodiation in the high-temperature ionization environment.101) Chen et al. also suggested that protonated carbohydrates were easy to decompose through the dehydration reaction. Cancilla et al. reported that the fragmentation behavior of carbohydrates was also related to their degree of branching.106) A branched glycan can establish multiple bindings to an alkali metal ion, resulting in lesser fragments. Computational methods, including MD simulation, semi-empirical and ab initio calculations, were utilized by Fukui et al. to investigate the relationship between the structure and reactivity (fragmentation) of sodiated oligosaccharides.105) Fukui et al. found that the coordination of metal ions to oligosaccharides affected the propensity for fragmentation. Based on the calculation result, increasing the number of oxygen atoms interacting with the sodium ion stabilized the carbohydrate structure (less fragmentation) and increased the ion yield.
Based on the understanding of mechanisms of MALDI and the nature of carbohydrates, three strategies are generally adopted to improve ion yield of carbohydrates: 1) thermalizing high-temperature ionization environments to reduce dealkalization and fragmentation; 2) doping ionization agents (permanent charges) to enhance ion yield; 3) changing the nature of glycans by derivatizations. We highlighted some of selected advancements in Table 1.
| Strategy | Methods | Ref. |
|---|---|---|
| Mediating high-temperature ionization environment | Soft ionization by ionic liquid matrices (ILMs) | |
| –1,1,3,3-Tetramethylguanidium (TMG) salt of p-coumaric acid (G3CA) | 107 | |
| –3-Aminoquinoline (3-AQ) based ILM | 108, 109 | |
| –2,4,6-Trihydroxyacetophene (THAP) based ILM | 110 | |
| Ice as a soft matrix | 111 | |
| Temperature-controlled sample plate allowing a frozen sample inside the vacuum | 112 | |
| Trylayer sample preparation method with thermal energy-dissipating diamond nanoparticles | 113 | |
| Doping ionization agent | Ammonium salts | 114, 115 |
| Introducing permanent charges with succinimidyloxycarbonylmethyl tris(2,4,6-trimethoxyphenyl) phosphonium bromide (TMPP-AcOSu) | 116 | |
| Nanoparticles containing ionization agents | 117–123 | |
| Chemical derivatization | Permethylation | 124, 127, 135 |
| Amidation | 126, 128, 132 | |
| Esterification | 133, 134 | |
| Derivatization with hydrazines | 125, 129 |
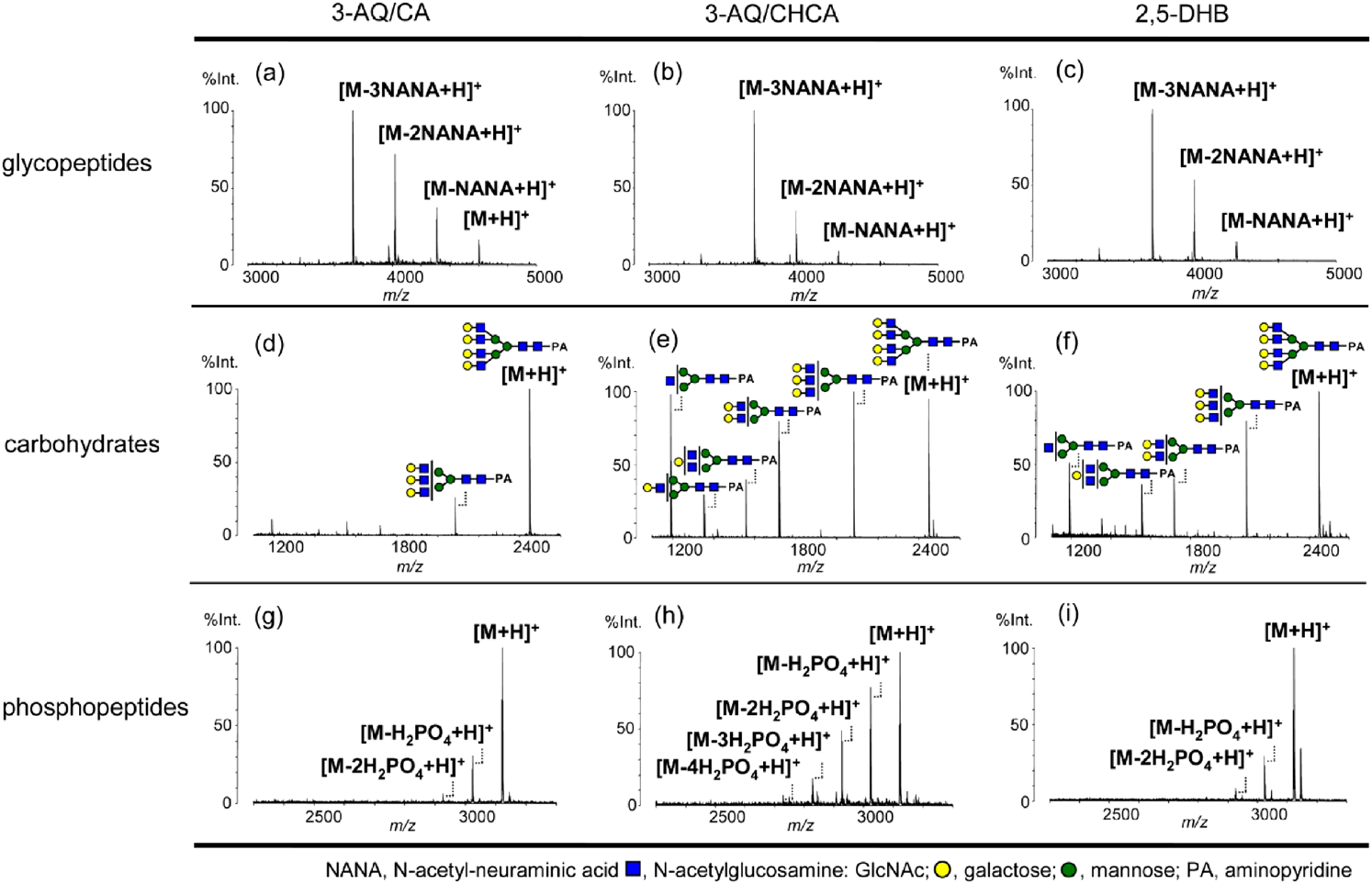
The selection of matrices is a crucial step for the analysis of carbohydrates because every matrix produces unique ionization environment after the laser irradiation.22) Problems associated with decomposition of thermally labile moieties and low ionization efficiency could be overcome by a suitable selection of the matrix. As shown in Fig. 3, “cool” ionic liquid matrices: 1,1,3,3-tetramethylguanidium (TMG) salt of p-coumaric acid (G3CA),107) and 3-aminoquinoline/p-coumaric acid (3-AQ/CA)108,109) were developed by Tanaka and coworkers to yield a less locally heated ionization environment for enhancing the sensitivity and suppressing the losses of sialic acid moieties. Applying a similar concept, THAP-based ionic liquid matrices110) were also reported for automated identification and quantitative analysis of native oligosaccharides. Witt et al. utilized ice as an extremely soft matrix to prevent carbohydrates from decomposition and enhance the capability of MALDI mass spectrometry for the structural characterization of glycans.111) The use of ice as a matrix is advantageous because it provides an ideal “near-physiological” condition. Liang et al. modified the ion source of commercial MALDI time-of-flight mass spectrometer to enable the preparation of a frozen carbohydrate/DHB sample at 100 K. Enhancement of sensitivity (20–30 folds) and reduction of fragmentation were achieved by this method.112) Our group developed a trilayer sample preparation method, which changes sample morphology by consecutively depositing matrices, diamond nanoparticles, and carbohydrates. Owing to the high thermal conductivity, diamond nanoparticles thermalized the MALDI temperature. Diamond nanoparticles also adjusted the incorporation of matrices and carbohydrates. As shown in Fig. 4, we selectively enhanced the sensitivity of underivatized carbohydrates by more than an order of magnitude from protein/carbohydrate mixtures.113) This sample preparation method is potentially suitable for diagnosing carbohydrates in real samples because carbohydrates are often accompanied by proteins in real biological samples.
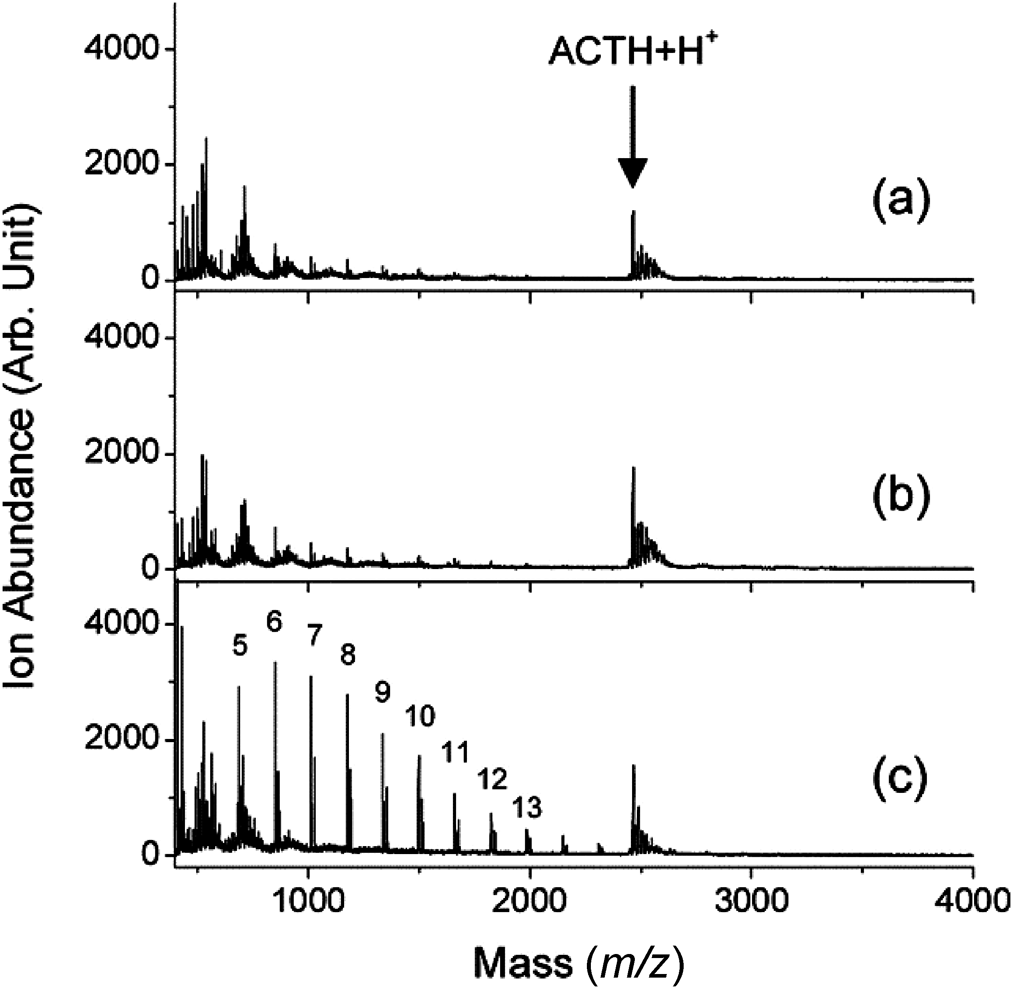
In the second category, Yamagaki et al. improved ionization efficiency of neutral glycans by adding ammonium salts in MALDI samples to form chloride-anionized molecules.114) In a similar fashion, doping salts containing anions, such as NO3− and Cl−, was conducted by Domann et al. to enhance the ionization efficiency of neutral N-linked glycans.115) Domann et al. reported the best matrix for ionizing glycans was mixing THAP with ammonium nitrate (1 M) to form [M+NO3]−. Gao et al. reported a derivatization method that enhanced 20- to 50-fold ion abundance of N-linked glycans by introducing permanent charges using nonspecific proteolysis with pronase E and co-derivatization with succinimidyloxycarbonylmethyl tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP-AcOSu) and methylamidation.116) Nanoparticles (NPs), such as gold, silver, titanium dioxide, iron oxide (Fe3O4), zinc oxide, and manganese oxide, have been utilized for the analysis of carbohydrates because they enhance ionization efficiency by releasing ionization agents.117–121) After absorbing laser energy, NPs also convert photon energy to thermal energy to facilitate material desorption. Charge and energy transfer from NPs to analytes triggers ionization/fragmentation and desorption of carbohydrates.117,122) Furthermore, functionalization of NPs with conventional organic matrices have been reported to serve as new matrix materials to preserve the intact glycan as well as the generation of extensive cleavages in MALDI.123) More details about methods on the structural identification of carbohydrates are reviewed in the following section.
Chemical derivatization has been accepted as a well-established option for the analysis of carbohydrates in MALDI124) because chemical derivatization protects glycans from losing acetyl groups or sialic acid moieties to increase carbohydrate ionization efficiency.125,126) Hakomori first demonstrated permethylation of complex carbohydrates.127) Derivatization was also reported to be beneficial to compensate differences in ionization efficiency. A novel amidation using acetohydrazide was demonstrated by Toyoda et al. to eliminate the discrepancy of ionization efficiency between α2,3- and α2,6-linked sialic acids-containing oligosaccharides.128) Derivatization also improves chromatographic analysis by reducing the polarity of glycans. Various methods for derivatization have been developed, such as uses of hydrazines129) and other labeling techniques.130)
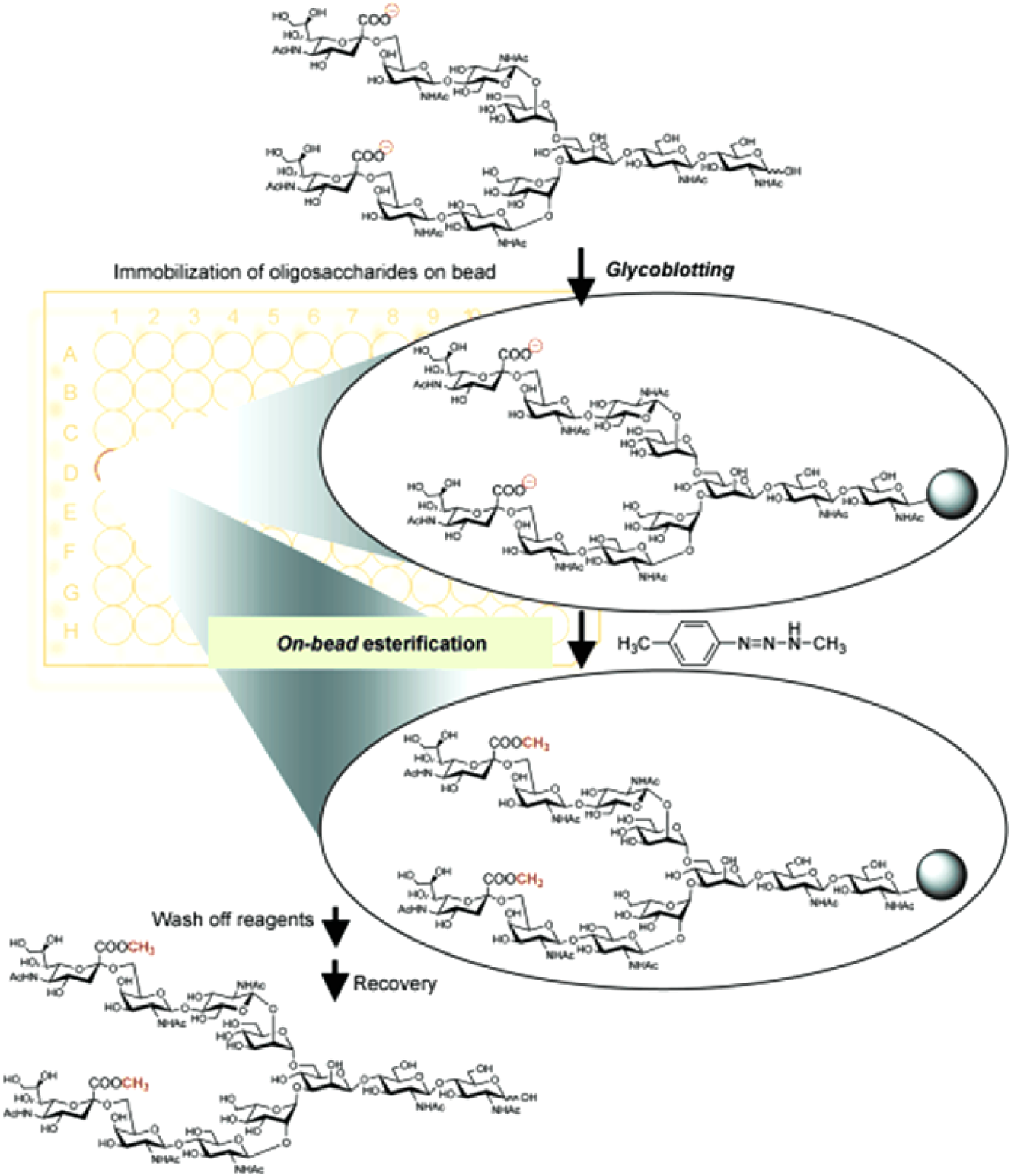
Although derivatization provides advantages, conventional derivatization typically requires harsh conditions (strong base) and long reaction times. Most derivatization methods require clean-up procedures for the removal of chemicals, resulting in a loss of samples, which especially leads to a serious problem for trace analysis.131) Thus, derivatization methods have been improved to meet the demand of low sample loss and high efficiency.132) Miura et al. reported a solid-phase methyl esterification of sialic acid residues of glycans for rapid and quantitative glycoform profiling by MALDI-TOF MS.133) As shown in Fig. 5, the workflow of their on-bead/on gold nanoparticle derivatization is potentially suitable for the large-scale glycomics study. Using a combination of carboxylic acid activators in ethanol, Reiding et al. achieved nearly complete ethyl esterification of α2,6-linked sialic acids and lactonization of α2,3-linked variants in a short time and mild conditions.134) A solid-phase miniaturized approach was demonstrated by Kang et al. for quantitative permethylation of oligosaccharides in less than a minute.135) By mixing analytes with methyl iodide in dimethyl sulfoxide solution, this method avoided excessive clean-up and yielded picomole-level sensitivity.
Structural information of carbohydrates, including sequence from glycosidic cleavages (C–O bonds) and branching and linkages from cross-ring cleavages (C–C bonds), is available by utilizing multiple fragmentation techniques in tandem mass spectrometry. Fragmentation techniques are essential methods to deposit energy into a targeted ion to cause reproducible bond cleavages. Cleavages yield diagnostic and interpretable fragment ions that reveal structural or sequence information about the molecule of interest. Commonly used fragmentation techniques in MALDI mass spectrometry include laser-induced promptly in-source decay (ISD),136) post-source decay (PSD, a unimolecular dissociation of metastable ions),137) and collision-induced dissociation (CID).138–140)
ISD ions are promptly produced by thermal activation and/or chemical reactions during desorption/ionization processes in the source region, such as hydrogen radical mediated141) and/or free electron mediated reactions.142,143) More energy input144) and higher energy-coupling efficiency between matrices and analytes89) enhance the ISD process. Because ISD ions receive the full acceleration energy in the ion source, they can be well resolved in the mass spectra. On the other hand, PSD ions are typically generated on the time scale of micro- to milliseconds in the field-free region. Owing to having the same velocity as their precursor ions, PSD ions cannot be resolved in linear TOF instruments.137) In order to resolve PSD ions, which are usually buried into the background in linear TOF instruments, it is necessary to analyze such ions by sweeping reflector potentials in reflectron TOF instruments. CID ions are generated through collisions of precursor ions with neutral gas molecules.145) During collision, redistribution of the kinetic energy of neutral gas molecules to the internal energy of precursor ions leads to fragmentation.145) A systematic nomenclature developed by Domon and Costello has become the common language for the community to specify fragmentation patterns of carbohydrates.146) Our goal for this section is to review some case studies of those fragmentation techniques in the structural analysis of carbohydrates. Readers are encouraged to refer to some of the more comprehensive reviews, which elucidate mechanisms of fragmentation techniques.89,137,141–143,147–151)
Complementary fragmentation methods were utilized to gain the structural information of carbohydrates. Suzuki et al. and Jovanović et al. conducted comprehensive structural identifications of neutral oligosaccharides in the negative ion mode utilizing ISD, PSD, and CID.152,153) Both groups reported that more information on linkage and branching, which were unobtainable using positive ion mode, could be acquired in the negative ion mode. Derivatization of carboxyl groups by methylamine using the condensing reagent (7-azabenzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluoro-phosphate (PyAOP) was applied by Nishikaze et al. to conduct glycoprofiling.154) They found that positive-ion CID of derivatized glycopeptides mainly provided information on peptide sequence and glycan composition, whereas negative-ion CID provided in-depth structural information on glycan moieties. PSD and CID were combined by Lattova et al. to profile derivatized N-linked glycans from ovalbumin.155) Compared to MALDI-PSD, they found that more structural information from cross-ring cleavages (a- and x-type fragments) could be obtained using CID in tandem mass spectrometry. Laser-induced ISD combined with high-energy collision-induced dissociation (heCID) was reported by Wuhrer et al. to be a capable method for targeted structural identification of glycans.150) Another design that involved two consecutive time-of-flight (TOF/TOF) mass analyzers combined with a collision cell and a timed ion selector was developed by Suckau and coworkers.156) MALDI TOF/TOF was reported to yield more cross-ring cleavages that allow us to access the information of linkage positions.157,158)
Akin to an ISD process, functionalized nanoparticles release charges and redistribute energy that facilitate fragmentation after absorbing laser energy.122) Our group synthesized DHB-functionalized mercury telluride nanoparticles (HgTe@DHB NPs) as a novel matrix, which induced both glycosidic and cross-ring cleavages in the solid phase. This method allowed us to do pseudo-MS/MS in a single step by regulating amounts of HgTe@DHB. Moreover, it preserved intact underivatized glycans. We have shown proof-of-concept for the novel matrix that permits the structural characterization of glycans.
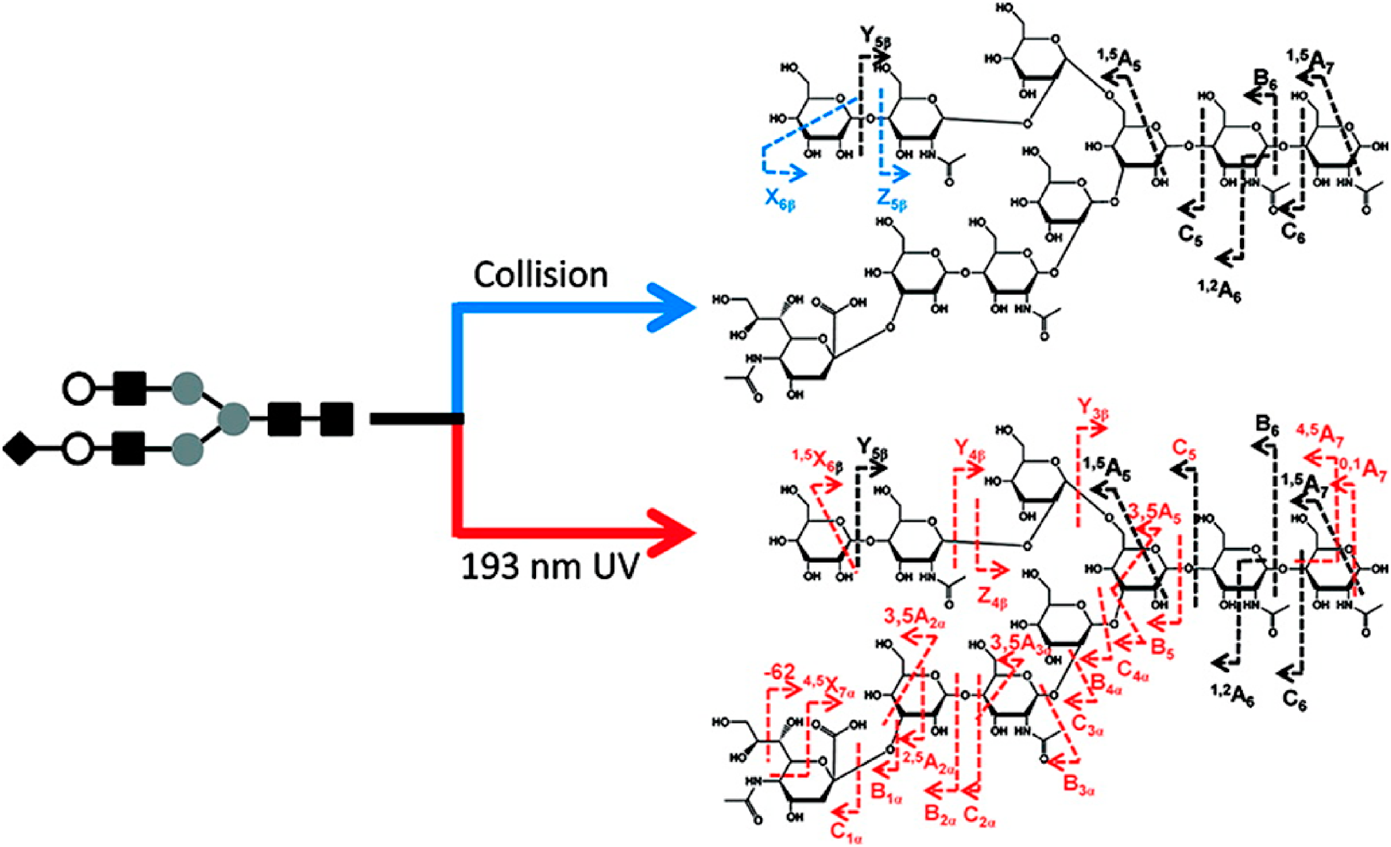
Dissociation could also be initiated by absorption of one or more photons (mainly ultraviolet photodissociation (UVPD) and infrared multiphoton dissociation (IRMPD)).159,160) Both UVPD and IRMPD were utilized for the structural analysis of carbohydrates in ion traps.161,162) As shown in Fig. 6, Ko et al. reported that, in contrast to CID, UVPD (193 nm) produced a more extensive series of cross-ring cleavage ions of deprotonated sialylated oligosaccharides, which yielded greater information for identification of glycans.162) Noticeably, no single fragmentation technique is yet able to acquire comprehensive structural information. Different fragmentation methods activate specific fragmentation channels. Complementary techniques have to be used in combination to obtain the complete structure.163,164)
Structural identification of carbohydrates is beneficial for gaining insights into cancer pathogenesis as well as evaluating the effectiveness of therapy. In this review, mechanisms of MALDI mass spectrometry, including ionization and desorption, are discussed. A deep understanding in MALDI mechanisms has enabled progress in its application to the analysis of carbohydrates. By developing novel matrices and sample preparation methods, the harsh ionization environment of conventional MALDI is mediated to facilitate the improvement of ion abundance. Ion yield is enhanced by doping ionization agents that contain permanent charges. Various derivatization methods, such as permethylation, amidation, esterification, and derivatization with hydrazines, are developed that reduce the neutral loss of thermally labile moieties and increase the detection sensitivity. Meanwhile, advances in ion activation, including photo- and collision-induced fragmentation techniques, yield remarkable structural details (sequencing, branching, and linkages) of carbohydrates. These developments benefit the field of glycomics. In many respects, carbohydrates are more challenging targets compared to others, such as proteins. More people from multidisciplinary fields are urged to work together for further developments to achieve comprehensive high-throughput structural identification of carbohydrates.