2021 Volume 62 Issue 1 Pages 29-35
2021 Volume 62 Issue 1 Pages 29-35
Amanita chuformis, a new species in the A. pseudogemmata-A. ballerina subclade of Amanita section Phalloideae, is described from China with both multi-gene phylogenetic and morphological data. This species occurs in subalpine coniferous forests in southwestern China and is characterized by its brownish pileus decorated with conical to patch-like volval remnants, a slightly striate pileal margin, a marginate basal bulb, and weakly amyloid to amyloid, subglobose to broadly ellipsoid basidiospores measuring 9.5–11 × 8–9.5 μm. Phylogenetic analyses based on internal transcribed spacer (ITS) region, the nuclear ribosomal RNA large subunit (nrLSU) and the genes for the polymerase II second largest subunit (RPB2) and for translation elongation factor 1-α (TEF1α) indicate A. chuformis is close to A. pseudogemmata and A. levistriata. The new species is described, illustrated and compared with closely related and similar species.
Amanita Pers. is a cosmopolitan genus comprising species with important economic, ecological and scientific values (Gilbert, 1940, 1941; Corner & Bas, 1962; Bas, 1969; Yang, 1997, 2005, 2015; Neville & Poumarat, 2004; Cui, Cai, Tang, Liu, & Yang, 2018). This genus contains not only edible mushrooms but also poisonous ones (Yang, 1997, 2005, 2015; Cai et al., 2014; Cai, Cui, & Yang, 2016; Cui et al., 2018). In addition, ca, 90% species in this genus can form ectomycorrhizal (ECM) relationships with a wide range of host plants, which are valuable for ecological protection and can be applied to study ECM biology (Yang, 2005, 2015; Cui et al., 2018).
Since the establishment of Amanita by Persoon in 1797, Amanita was historically split into several genera or various subdivisions (Veselý, 1933; Gilbert, 1941; Konrad & Maublanc, 1948; Singer, 1951, 1986; Garcin, 1984). Corner & Bas (1962) and Bas (1969) proposed to split Amanita into two subgenera and six sections, which was later revised as two subgenera and seven sections by Yang (1997) and widely accepted by many mycologists (Weiß, Yang, & Oberwinkler, 1998; Drehmel, Moncalvo, & Vilgalys, 1999; Oda, Tanaka, & Tsuda, 1999; Zhang, Yang, & Yang, 2004). Vizzini et al. (2012) split Amanita subsection Vittadiniae Bas (1969) emend. Tulloss et al. (2016) as a separate genus, namely Aspidella E.-J. Gilbert, and then Redhead et al. (2016) revised its name as Saproamanita Redhead, Vizzini, Drehmel & Contu. However,Tulloss et al. (2016) disagreed with this generic splitting. According to multi-gene phylogenetic and morphological data, Cui et al. (2018) treated Amanita subsection Vittadiniae Bas (1969) emend. Tulloss et al. (2016) as a member of Amanita and divided the genus into three subgenera and eleven sections, namely section Amanita, section Amarrendiae (Bougher & T. Lebel) Zhu L. Yang, Y.Y. Cui, Q. Cai & L.P. Tang, section Caesareae Singer ex Singer and section Vaginatae (Fr.) Quél. in subgenus Amanita; section Amidella (E.-J. Gilbert) Konrad & Maubl., section Arenariae Zhu L. Yang, Y.Y. Cui & Q. Cai, section Phalloideae (Fr.) Quél., section Roanokenses Singer ex Singer, section Strobiliformes Singer ex Q. Cai, Zhu L. Yang & Y.Y. Cui and section Validae (Fr.) Quél. in subgenus Amanitina (E. -J. Gilbert) E. -J. Gilbert; section Lepidella Corner & Bas in subgenus Lepidella Beauseigneur. Based upon the multi-gene phylogenetic analyses in Cui et al. (2018), Amanita section Phalloideae could be divided into three subclades, i.e, lethal Amanita subclade, A. pseudogemmata-A. ballerina subclade, and A. hesleri-A. zangii subclade. These three subclades can be separated with both molecular and morphological data and may represent different subsections in section Phalloideae. However, no more taxonomic arrangements have been done because few species were known from the latter two subclades.
During the study of agarics in southwestern China, we have collected two interesting specimens representing a member of A. pseudogemmata-A. ballerina subclade. Subsequent morphological and molecular analyses with a multi-gene dataset confirmed that they are indeed a taxon in the subclade and can be separated from the others. Therefore, a new species is described herein.
The examined specimens were collected during the years of 2014–2017 from subalpine forests in Medog County, Tibet Autonomous Prefecture, China, and are deposited in the Herbarium of Cryptogams, Kunming Institute of Botany, Chinese Academy of Sciences (HKAS). Macroscopic descriptions are based on detailed field notes and digital images. The color codes of the form “4C2” indicate the plate, row and color block from Kornerup & Wanscher (1981). For microscopic studies, free-hand sections of dried basidiomata were prepared. The ratio of the length of striation to the radius of the pileus is suffixed with R. Microscopic characters were observed on dried specimens mounted on 5% KOH, and stained in Congo red when necessary. Melzer’s reagent was used to test the amyloidity of basidiospores. The term “[n/m/p]” represents n basidiospores measured from m basidiomata of p collections. Dimensions for basidiospores are given using notation of the form (a–)b–c(–d). The range b–c contains a minimum of 90% of the measured values. Extreme values, a and d, are provided in parentheses. Q means the length/width ratio of a basidiospore in side view; Qm indicates average Q of all basidiospores measured ± sample standard deviation. SigmaPlot 10.0 (Systat Software, San Jose, California) was applied to calculate these values.
Methods for DNA extraction, PCR amplification, and sequencing protocols followed those of Cai et al. (2014, 2016), Liu et al. (2017),Cui et al. (2018) and references therein. Sequences of internal transcribed spacer (ITS) region, the nuclear ribosomal RNA large subunit (nrLSU) and the genes for the polymerase II second largest subunit (RPB2) and for translation elongation factor 1-α (TEF1α) were used for phylogenetic analyses. The universal primers ITS1F/ITS4 (White et al., 1990; Gardes & Bruns, 1993), LR0R/LR5 (Vilgalys & Hester, 1990), Am-6F/Am-7R (Cai et al. 2014, 2016; Cui et al., 2018) and 983F/1567R (Rehner & Buckley, 2005) were used for the amplification and sequencing of ITS, nrLSU, RPB2 and TEF1α, respectively. Detailed information about the sequences newly generated in this study and retrieved from GenBank can be found in Supplementary Table S1.
The sequences of four gene regions were aligned with MAFFT v. 7.310 (Katoh & Standley, 2013) and manually checked with Bioedit v. 7.0.9 (Hall, 1999) separately. For ITS, Gblocks v. 0.91b (Castresana, 2000) was used to detect and exclude the ambiguously aligned regions, with options “Allow smaller final blocks” and “Allow gap positions within the final blocks”. Separate single-gene analyses were conducted to examine the conflict among topologies with maximum likelihood (ML). Phyutility v. 2.2 (Smith & Dunn, 2008) was then applied to concatenate the four gene regions. The concatenated alignment was deposited in TreeBASE (http://www.treebase.org/treebase/) with submission ID 26435. MrModeltest v. 2.3 (Nylander, 2004) was used to choose the most appropriate substitution model for each dataset under Akaike information criterion (AIC). Maximum Likelihood (ML) and Bayesian Inference (BI) were used for phylogenetic analyses based on RAxML v. 7.2.6 (Stamatakis, 2006) and MrBayes v. 3.1.2 (Ronquist & Huelsenbeck, 2003), respectively. In the multi-gene analyses, representive species of section Arenariae [A. arenaria (O.K. Mill. & E. Horak) Justo], section Validae (A. citrinoannulata Y.Y. Cui, Q. Cai & Zhu L. Yang) and section Strobiliformes (A. aspericeps Y.Y. Cui, Q. Cai & Zhu L. Yang) were selected as outgroup because these three sections are close to sect. Phalloideae based on the study of Cui et al. (2018). For the ML analyses, statistical supports for internodes were calculated using nonparametric bootstrapping with 1,000 replicates (ML bootstrap: MLB). BI analyses were conducted with generations set to 2 million, and trees sampled every 100 generations. The convergence was judged with the average standard deviation of split frequencies (< 0.01) and the effective sample size (ESS) values (> 200). The sampled trees were summarized after omitting the first 25% of trees as burn-in by using the “sump” and “sumt” commands, and the Bayesian posterior probabilities (BPP) of clades were estimated based on the majority rule consensus with the remaining trees.
No obvious differences in topology were observed when the four genes were analyzed individually (Supplementary Figs S1-S2-S3-S4). The GTR+GAMMA+I model was the most appropriate substitution model for nrLSU and RPB2, while the GTR+GAMMA and SYM+GAMMA+I models were the best ones for ITS and TEF1α, respectively. In the multi-gene dataset, a total of 160 sequences, including 7 newly generated and 153 retrieved from GenBank, were assembled from four genes (47 for ITS, 44 for nrLSU, 34 for TEF1α and 35 for RPB2). The aligned multi-gene dataset contained 2,600 nucleotides (472 for ITS, 867 for nrLSU, 583 for TEF1α and 678 for RPB2). Topologies of phylogenetic trees generated from ML and BI analyses were almost identical with minimal variation in statistical supports, and thus only the tree inferred from ML analysis was displayed (Fig. 1; Supplementary Fig S1 S2 S3 S4).

Taxonomy
Amanita chuformis Yang-Yang Cui, Qing Cai & Zhu L. Yang, sp. nov., Figs. 1 2 3.
MycoBank no.: MB 835323.
Diagnosis: Amanita chuformis is characterized by its greyish to brownish pileus covered with greyish to brownish subconical to patch-like volval remnants, a slightly striate pileal margin, a marginate basal bulb, weakly amyloid to amyloid, subglobose to broadly ellipsoid basidiospores (9.5–11 × 8.0–9.5 μm) and absence of clamps. In addition, this species is currently only collected in subalpine forests with trees of Abies and Picea.

Type: CHINA, Tibet Autonomous Prefecture, Medog County, in subalpine forests with trees of Abies and Picea, altitude 3217 m, 26 Jul 2017, Si-Peng Jian 39 (HKAS 101028, holotype, GenBank accession No.: ITS = MT395379, nrLSU = MT395381, TEF1α = MT364257, RPB2 = MT364258).
Etymology: “chuformis” refers to the shape of basal bulb similar to the traditional Chinese tool “chu”.
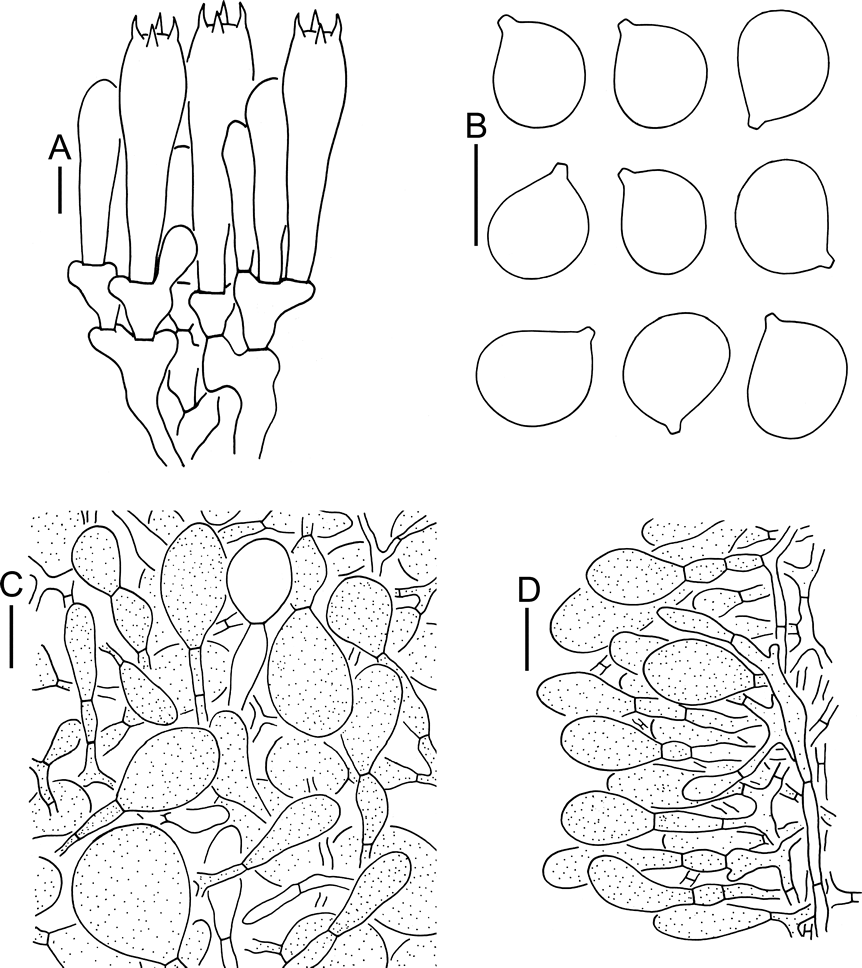
Basidioma medium-sized. Pileus 5–8 cm diam, convex-applanate to applanate, without an umbo or depression at center, brownish gray (4C2–4, 3C2–4), brownish (4B2–3, 2C2–4) to dirty white (2B2–3), covered with grayish black (3E2–4), grayish brown (4D2–5) to brownish (4B2–3, 2C2–4), pyramidal, conical to subconical volval remnants, often stick together as flat patches; margin slightly striate (0.07–0.1 R) and non-appendiculate; trama white (1A1), unchanging. Lamellae free, crowded, white (1A1); lamellulae truncate, plentiful. Stipe 10–12.5 cm long, 0.5–1.2 cm diam, subcylindric and slightly tapering upwards, with apex slightly expanded, grayish (1C2–4), brownish (4C2–5, 4B2–3, 2C2–4) to dirty white (2B2–3), covered with grayish (1C2–4) to brownish (4C2–5, 4B2–3, 2C2–4) fibrils; context white (1A1), unchanging; basal bulb marginate, dirty white (2B2–3) to white (1A1), upper edge with shortly limbate, grayish (1C2–4) to brownish (4C2–5, 4B2–3, 2C2–4) volval remnants. Annulus apical, membranous, white to dirty white (2B2–3) at upper surface, grayish (2B1–3, 1C2–4) to gray (2D2–4) at lower surface, gray (2D2–4) to brownish gray (4D2–4, 4E2–4) at lamellar edge. Odor not distinctive.
Lamellar trama bilateral. Mediostratum 25–40 μm wide, composed of abundant ellipsoid to clavate inflated cells (60–150 × 20–40 μm); filamentous hyphae abundant, 2–8 μm wide; vascular hyphae scarce. Lateral stratum composed of abundant ellipsoid to clavate inflated cells (40–100 × 10–25 μm), diverging at an angle of ca, 30 to 45° to mediostratum; filamentous hyphae abundant and 3–7 μm wide. Subhymenium 25–45 μm thick, with 2–3 layers of subglobose, ovoid to ellipsoid or irregular cells, 10–20 × 10–20 μm. Basidia 50–65 × 10–15 μm, clavate, 4-spored; sterigmata 3–6 μm long; basal septa without clamps. Basidiospores [131/5/2] (9–)9.5–11(–12) × 8–9.5(–10.5) μm, Q = (1–)1.05–1.29(–1.32), Qm = 1.16 ± 0.06, subglobose to broadly ellipsoid, weakly amyloid to amyloid, colorless, thin-walled, smooth; apiculus small (ca, 1–1.5 μm in width and length). Lamellar edge appearing as a sterile strip, composed of globose, subglobose, fusiform to elongate or sphaeropedunculate inflated cells (20–85 × 10–50 μm), single and terminal or in chains of 2–3, thin-walled, colorless; filamentous hyphae abundant, 3–9 μm wide, irregularly arranged or running parallel to lamellar edge. Pileipellis 70–120 μm thick; upper layer (30–55 μm thick) non- or slightly gelatinized, composed of subradially to somewhat interwoven, thin-walled, colorless, filamentous hyphae 2–6 μm wide; lower layer (40–65 μm thick) composed of radially and compactly arranged, colorless, filamentous hyphae 2–10 μm wide; vascular hyphae scarce. Volval remnants on pileus composed of irregularly arranged elements: filamentous hyphae rare to scattered, 2–8 μm wide, colorless to yellow-brown, thin-walled, branching, anastomosing; inflated cells very abundant to nearly dominant, globose, subglobose, fusiform to ellipsoid, 15–60 × 10–55 μm, yellow-brown, thin-walled, terminal or in chains of 2–3; vascular hyphae scarce. Outer layer of volval limb on the stipe base composed of abundant to very abundant, subglobose, fusiform, ellipsoid to short clavate, yellow-brown inflated cells (20–95 × 10–60 μm), mixed with scattered yellow-brown filamentous hyphae (2–5 μm wide), which becoming fairly abundant to abundant towards inner layer. Stipe trama composed of longitudinally arranged, long clavate, terminal cells, 75–350 × 20–40 μm; filamentous hyphae scattered to abundant, 3–15 μm wide; vascular hyphae scarce. Interior of annulus composed of very abundant to dominant filamentous hyphae (2–5 μm wide), mixed with rare globose, subglobose to ellipsoid inflated cells (10–25 × 8–20 μm). Clamps absent in all parts of basidioma.
Habitat: Solitary to scattered in subalpine forests; basidioma occurring in summer and autumn.
Distribution: Known from southwestern China.
Additional specimen examined: CHINA, Tibet Autonomous Prefecture, Medog County, in subalpine forests with trees of Abies, altitude ca, 4000 m, 3 Aug 2014, Bang Feng 1689 (HKAS 94075, GenBank accession No.: ITS = MT395378, nrLSU = MT395380, TEF1α = MT364256).
Notes: The analysis in Cui et al. (2018) indicated that section Phalloideae could be divided into three subclades, which is consistent with the present study (Fig. 1). Unfortunately, the relationships among these three subclades are not well resolved and need further investigations (Fig. 1). Our multi-gene phylogenetic analyses strongly supported the monophyly of A. chuformis (MLB/BPP = 100/1) (Fig. 1). The new taxon is phylogenetically located in the A. pseudogemmata-A. ballerina subclade (Fig. 1), and its striate pileal margin, marginate basal bulb and weakly amyloid basidiospores match well with the common characters of this subclade. This subclade may be treated as an independent subsection or even a section until more species are recognized. To date, only five known species (A. ballerina Raspé, Thongbai & K.D. Hyde, A. chuformis, A. franzii Zhu L. Yang, Y.Y. Cui & Q. Cai, A. levistriata D.T. Jenkins and A. pseudogemmata Hongo) are recorded in this subclade, with molecular supports, and three of them (A. chuformis, A. franzii and A. pseudogemmata) are known from China (Fig. 1).
In our multi-gene phylogenetic analyses, A. pseudogemmata is close to A. levistriata, together they form a monophyletic group sister to A. chuformis (Fig. 1). However, A. levistriata, a species originally described from USA, differs from A. chuformis by its smaller basidioma with a ocher yellow to yellow pileus ca, 1.7–3 cm diam, a whitish to pale yellow stipe with a white to yellow, superior annulus, relatively smaller basidiospores (7.8–9.3 × 6.3–7.8 μm), and its association with mixed coniferous and deciduous trees on sandy soil (Jenkins, 1988; Tulloss & Yang, 2020). Amanita pseudogemmata, originally described from Japan and also known from China, can be distinguished from A. chuformis by its dirty yellow to yellow-brown basidioma, a white to yellowish, superior annulus and smaller basidiospores (7–9.5 × 6–8.5 μm) (Hongo, 1974; Doi, 1991; Yang & Doi, 1999; Yang, 2005, 2015; Cui et al., 2018). Furthermore, A. pseudogemmata is distributed in subtropical broad-leaved forests with trees of Fagaceae (Hongo, 1974; Doi, 1991; Yang & Doi, 1999; Yang, 2005; 2015; Cui et al., 2018). Within A. pseudogemmata-A. ballerina subclade, A. chuformis, A. levistriata and A. pseudogemmata form a monophyletic group with high supports (MLB/BPP = 100/1), which is related to A. franzii and A. ballerina (Fig. 1). Furthermore, A. franzii can be confused with A. chuformis in their similar color of basidiomata and same shape of basal bulbs. However, A. franzii has a superior annulus and relatively narrower basidiospores (8.5–10.5 × 6.5–7.5 μm, Q = 1.12–1.46, Qm = 1.34 ± 0.11) (Yang, 2015; Cui et al., 2018). In addition, A. franzii is distributed in subtropical broad-leaved or mixed forests with relatively lower altitudes between 900 m to 2000 m, compared with A. chuformis (Yang, 2015; Cui et al., 2018). Amanita ballerina, originally described from Thailand, differs from A. chuformis by its relatively smaller and dull white basidioma with a pileus ca, 3.4–4.2 cm diam, white to pale yellowish white volval remnants on the pileus firstly as a cottony layer and later often breaking into squamules or patches, relatively smaller basidiospores (7.5–8.9 × 6–7.5 μm), and distribution in evergreen Fagaceae hill forest or mixed deciduous Dipterocarpaceae/Fagaceae forest with altitudes between 740 m to 1170 m (Thongbai et al., 2017).
Morphologically, the marginate basal bulb of A. chuformis also reminds to the species in Amanita subsection Limbatulae Bas, including A. mutabilis Beardslee, A. sublutea (Cleland) E.-J. Gilbert and A. kammala Grgur. [also recorded as Amanita sp. indet. in Bas (1969) (described in page 539, illustrated in Figs. 333 and 334 in page 537)]. However, the American A. mutabilis has a pale tan to tannish cream pileus covered concolorous to slightly darker to nearly black patches, a non-striate pileal margin, white context with pink color change when cut, and ellipsoid to elongate basidiospores (11–13.5 × 6–8.5 μm, Q = 1.4–2.2) (Beardslee, 1919; Bas, 1969; Tulloss, 1984). Amanita sublutea and A. kammala, two species described from South Australia, share smaller basidiomata with pilei 3–4.5 cm diam, non-striate pileal margins, exannulate stipes, and elongate to cylindrical basidiospores (11.5–13 × 6.5–7.5 μm, Q = 1.65–2.05 for A. sublutea; 11.5–13.5 × 5.5–6.5 μm, Q = 2–2.4 for A. kammala) (Cleland, 1931; Gilbert, 1940; Bas, 1969; Grgurinovic, 1998).
Disclosure
The authors declare no conflicts of interest for this study. All the experiments undertaken in this study comply with the current laws of the People’s Republic of China.
We are grateful to Mr. Si-Peng Jian and Dr. Bang Feng (Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China) for providing valuable collections, and Dr. Jing Li (Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China) for her kind help in molecular analyses. Thanks are also due to the anonymous reviewers for their professional suggestions and comments. This work is supported by the biodiversity investigation, observation and assessment program (2019–2023) of Ministry of Ecology and Environment of China, and Yunnan Ten-Thousand-Talents Plan - Yunling Scholar Project.