2021 Volume 62 Issue 1 Pages 36-41
2021 Volume 62 Issue 1 Pages 36-41
Two new species, Hyphoderma fissuratum and H. mopanshanense spp. nov., are proposed based on morphological and molecular evidences. Hyphoderma fissuratum is characterized by resupinate basidiomata with cracking hymenial surface, a monomitic hyphal system with generative hyphae bearing clamp connections, IKI–, CB–, and cylindrical, colorless, thin-walled, smooth basidiospores measuring (8.5–10.3 × 3–4 µm). Hyphoderma mopanshanense is characterized by an annual growth habit, slight gray to cream hymenial surface, and fusiform, thick-walled cystidia apically encrusted with crystal. Sequences of ITS and LSU nrRNA gene regions of the studied samples were generated, and phylogenetic analyses were performed with maximum likelihood, maximum parsimony and Bayesian inference methods. These phylogenetic analyses based on molecular data of ITS and nLSU sequences showed that two Hyphoderma new species formed a well supported monophyletic lineage distinct from other related species and then H. fissuratum grouped with H. medioburiense and H. roseocremeum. Hyphoderma mopanshanense grouped with H. setigerum.
Hyphoderma Wallr. was typified by H. setigerum (Fr.) Donk (synonymy with H. spiculosum Wallr.) (Wallroth, 1833). The genus is characterized by resupinate to effuse-reflexed basidiomata with ceraceous consistencey, and smooth to tuberculate or hydnoid hymenophore and a monomitic hyphal structure (rarely dimitic) with clamp connections on generative hyphae, presence of cystidia or not, basidia suburniform to subcylindrial and cylindrical, ellipsoid to subglobose, smooth, thin-walled basidiospores (Wallroth, 1833; Bernicchia & Gorjón, 2010). So far 100 species have been accepted in the genus worldwide (Wallroth, 1833; http://www.indexfungorum.org/names/Names.asp; http://www.mycobank.org/Biolomics.aspx?Table=Mycobank).
Recently, molecular studies involving Hyphoderma based on single-gene or multi-gene datasets have been carried out (Larsson, 2007; Telleria, Dueñas, Beltrán-Tejera, Rodríguez-Armas & Martín, 2012; Binder et al., 2013; Yurchenko & Wu, 2014a, 2015; Justo et al., 2017). On the basis of the internal transcribed spacer (ITS) regions and the large subunit nuclear ribosomal RNA gene (nLSU) sequences, Larsson (2007) revealed the classification of corticioid fungi and showed that Hyphoderma obtusum J. Erikss. and H. setigerum (Fr.) Donk clustered into Meruliaceae Rea and then grouped with Hypochnicium polonense (Bres.) Å. Strid. Telleria, Dueñas, Beltrán-Tejera, Rodríguez-Armas & Martín. (2012) proposed a new species, Hyphoderma macaronesicum Tellería, M. Dueñas, Beltrán-Tej, Rodr.-Armas & M.P. Martín and then discussed the relationships with the closely related taxa in Hyphoderma.Binder et al. (2013) presented the molecular studies employing multi-gene datasets (5.8S, nLSU, translation elongation factor 1-α (TEF1) gene, mitochondrial rRNA gene sequences (mtSSU), the second-largest subunit of RNA polymerase II (RPB2) and the largest subunit of RNA polymerase II) to investigate the phylogenetic relationships within the Polyporales, in which Hyphoderma cremeoalbum (Höhn. & Litsch.) Jülich and H. setigerum were nested into the residual polyporoid clade and placed inside of Hyphodermataceae Jülich. Yurchenko & Wu (2014a) studied the Hyphoderma setigerum complex and showed that H. pinicola Yurchenko & Sheng H. Wu represented a fifth species of H. setigerum complex. Yurchenko & Wu (2015) reported that Hyphoderma moniliforme (P.H.B. Talbot) Manjón, G. Moreno & Hjortstam and H. nemorale K.H. Larss., saprobically growing on wood, were recorded as new for mycobiota in China. A revised family-level classification of the Polyporales revealed that four Hyphoderma species nested into the residual polyporoid clade belonging to Hyphodermataceae and then grouped with three genera Meripilus P. Karst., Physisporinus P. Karst. and Rigidoporus Murrill (Justo et al., 2017).
Recently, we collected two undescribed taxa from Yunnan Province, P. R. China that could not be assigned to any described genera. We present morphological and molecular phylogenetic evidence that support the recognition of two new species within the Hyphoderma, based on the internal transcribed spacer (ITS) regions and the large subunit nuclear ribosomal RNA gene (nLSU) sequences.
The specimens studied are deposited at the herbarium of Southwest Forestry University (SWFC), Kunming, Yunnan Province, P.R. China. Macromorphological descriptions are based on field notes. Color terms follow Petersen (1996). Micromorphological data were obtained from the dried specimens, and observed under a light microscope following Dai (2012). The following abbreviations were used: KOH = 5% potassium hydroxide, CB = Cotton Blue, CB– = acyanophilous, IKI = Melzer’s reagent, IKI– = both inamyloid and indextrinoid, L = mean spore length (arithmetic average for all spores), W = mean spore width (arithmetic average for all spores), Q = variation in the L/W ratios between the specimens studied, n (a/b) = number of spores (a) measured from given number (b) of specimens.
2.2 Molecular phylogenyCTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd, Beijing) was used to obtain genomic DNA from dried specimens, according to the manufacturer’s instructions with some modifications that a small piece of dried fungal specimen (about 30 mg) was ground to powder with liquid nitrogen. The powder was transferred to a 1.5 mL centrifuge tube, suspended in 0.4 mL of lysis buffer, and incubated at 65 °C in a water bath for 60 min. After that, 0.4 mL phenol-chloroform (24:1) was added to each tube and the suspension was shaken vigorously. After centrifugation at 13,000 rpm for 5 min, 0.3 mL of supernatant was transferred to a new tube and mixed with 0.45 mL of binding buffer. The mixture was then transferred to an adsorbing column (AC) for centrifugation at 13,000 rpm for 0.5 min. Then, 0.5 mL of inhibitor removal fluid was added in AC for a centrifugation at 12,000 rpm for 0.5 min. After washing twice with 0.5 mL of washing buffer, the AC was transferred to a clean centrifuge tube, and 100 mL elution buffer was added to the middle of adsorbed film to elute the genome DNA. ITS region was amplified with primer pair ITS5 and ITS4 (White, Bruns, Lee, & Taylor, 1990). Nuclear LSU region was amplified with primer pair LR0R and LR7 (http://www.biology.duke.edu/fungi/mycolab/primers.htm). The PCR procedure for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 58 °C for 45 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94 °C for 1 min, followed by 35 cycles at 94 °C for 30 s, 48 °C 1 min and 72 °C for 1.5 min, and a final extension of 72 °C for 10 min. The PCR products were purified and directly sequenced at Kunming Tsingke Biological Technology Limited Company. All newly generated sequences were deposited at GenBank (Table 1).
Species name |
Sample no. |
GenBank accession no. |
References |
||
ITS |
nLSU |
||||
Hyphoderma cremeoalbum |
NH 11538 |
DQ677492 |
DQ677492 |
||
H. definitum |
GEL 2898 |
— |
AY515348 |
Yurchenko & Wu (2014a) |
|
H. definitum |
FCUG 2426 |
AJ534293 |
— |
||
H. definitum |
NH 12266 |
DQ677493 |
DQ677493 |
||
H. fissuratum |
CLZhao 6726 |
MT791330 |
MT791334 |
Present study |
|
H. fissuratum |
CLZhao 6731 |
MT791331 |
MT791335 |
Present study |
|
H. granuliferum |
KHL 12561 |
JN710545 |
JN710545 |
Yurchenko & Wu (2014b) |
|
H. incrustatum |
KHL 6685 |
— |
AY586668 |
Yurchenko & Wu (2014b) |
|
H. litschaueri |
NH 7603 |
DQ677496 |
DQ677496 |
||
H. litschaueri |
CFMR 2011050 |
KJ140573 |
— |
Yurchenko & Wu (2014b) |
|
H. macaronesicum |
TFC 15981 |
HE577027 |
— |
Yurchenko & Wu (2014b) |
|
H. macaronesicum |
MA 16099 |
HE577028 |
— |
Yurchenko & Wu (2014b) |
|
H. medioburiense |
NH 10950 |
DQ677497 |
DQ677497 |
||
H. moniliforme |
TNM F14735 |
KC928282 |
KC928283 |
Yurchenko & Wu (2015) |
|
H. moniliforme |
TNM F14739 |
KC928284 |
KC928285 |
Yurchenko & Wu (2015) |
|
H. mopanshanense |
CLZhao 6493 |
MT791328 |
MT791332 |
Present study |
|
H. mopanshanense |
CLZhao 6498 |
MT791329 |
MT791333 |
Present study |
|
H. nemorale |
EM 2793 |
— |
AY586669 |
Yurchenko & Wu (2014b) |
|
H. nemorale |
TNM F3910 |
KC928282 |
KC928283 |
Yurchenko & Wu (2015) |
|
H. nemorale |
TNM F3931 |
KJ885183 |
KJ885184 |
Yurchenko & Wu (2015) |
|
H. nudicephalum |
Wu 9508225 |
AJ534268 |
— |
||
H. nudicephalum |
Wu 930729 |
AJ534269 |
— |
||
H. nudicephalum |
GEL 4727 |
— |
AJ406510 |
Yurchenko & Wu (2014b) |
|
H. obtusiforme |
KHL 1464 |
JN572909 |
— |
Yurchenko & Wu (2014b) |
|
H. obtusiforme |
KHL 11105 |
JN572910 |
— |
Yurchenko & Wu (2014b) |
|
H. obtusum |
JS 17804 |
— |
AY586670 |
Yurchenko & Wu (2014b) |
|
H. occidentale |
KHL 8469 |
— |
AY586674 |
Yurchenko & Wu (2014b) |
|
H. occidentale |
KHL 8477 |
DQ677499 |
DQ677499 |
||
H. pinicola |
TNM F13635 |
KJ885179 |
KJ885180 |
Yurchenko & Wu (2014b) |
|
H. pinicola |
TNM F13643 |
KC928278 |
KC928279 |
Yurchenko & Wu (2014b) |
|
H. prosopidis |
ARIZ 8479 |
HE577029 |
— |
Yurchenko & Wu (2015) |
|
H. roseocremeum |
NH 10545 |
— |
AY586672 |
Yurchenko & Wu (2014b) |
|
H. setigerum |
FCUG 1688 |
AJ534272 |
— |
||
H. setigerum |
FCUG 1200 |
AJ534273 |
— |
||
H. setigerum |
KHL 8544 |
— |
AY586673 |
Yurchenko & Wu (2014b) |
|
H. setigerum |
NH 8544 |
— |
FN907905 |
Yurchenko & Wu (2014b) |
|
H. subtestaceum |
HHB 11620 |
GQ409521 |
— |
Yurchenko & Wu (2014b) |
|
H. subtestaceum |
MJL 1536 |
GQ409522 |
— |
Yurchenko & Wu (2014b) |
|
H. subtestaceum |
Wu 920215 |
AJ534278 |
— |
||
H. subtestaceum |
Wu 930418 |
AJ534277 |
— |
||
H. transiens |
NH 12304 |
DQ677504 |
DQ677504 |
||
Phanerochaete sordida |
KHL 12054 |
EU118653 |
EU118653 |
||
Sequences were aligned in MAFFT 7 (https://mafft.cbrc.jp/alignment/server/) using the “G-INS-I” strategy for nLSU, and manually adjusted in BioEdit (Hall, 1999). Alignment datasets were deposited in TreeBase (submission ID 25203). Phanerochaete sordida (P. Karst.) J. Erikss. & Ryvarden was selected as outgroup for phylogenetic analyses of ITS and nLSU phylogenetic trees (Yurchenko & Wu, 2015).
Maximum parsimony analyses were applied to the ITS and nLSU dataset sequences. Approaches to phylogenetic analysis followed Chen, Cui, and Dai (2016), and the tree construction procedure was performed in PAUP* version 4.0b10 (Swofford, 2002). All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap (BT) analysis with 1,000 replicates (Felsenstein, 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each Maximum Parsimonious Tree generated. Datamatrix was also analyzed using Maximum Likelihood (ML) approach with RAxML-HPC2 through the Cipres Science Gateway (www.phylo.org; ) (Miller et al., 2009). Branch support (BS) for ML analysis was determined by 1000 bootstrap replicates.
MrModeltest 2.3 (Nylander, 2004) was used to determine the best-fit evolution model for each data set for Bayesian inference (BI). BI was calculated with MrBayes3.1.2 with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist & Huelsenbeck, 2003). Four Markov chains were run for 2 runs from random starting trees for 400,000 generations for ITS (Fig. 1), 300,000 generations for nLSU (Fig. 2), and trees were sampled every 100 generations. The first one-fourth generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated. Branches were considered as significantly supported if they received maximum likelihood bootstrap (BS)>70%, maximum parsimony bootstrap (BT) >50%, or Bayesian posterior probabilities (BPP) >0.95.

Hyphoderma fissuratum C.L. Zhao & X. Ma, sp. nov. Fig. 3, 4.
MycoBank no.: MB 836259.


Holotype: China, Yunnan Province, Chuxiong, Zixishan Forestry Park, E 101°24′, N 25°0′, alt. 1950 m, on angiosperm stump, 26 Jun 2018, CLZhao 6726 (SWFC).
Etymology: Fissuratum (Lat.): referring to the cracking hymenial surface.
Basidiomata: Annual, resupinate, thin, ceraceous when fresh, turn to leathery upon drying, up to 12 cm long, 5 cm wide, 300–500 µm thick. Hymenial surface smooth, cream when fresh, cracking, cream to cinnamon-buff upon drying. Sterile margin distinct, white.
Hyphal structure: Hyphal system monomitic, generative hyphae with clamp connections, IKI–, CB–, tissues unchanged in KOH.
Subiculum: Generative hyphae colorless, thin-walled, rarely branched, 3.5–5.5 μm diam; larger crystal present.
Hymenial layer: Generative hyphae colorless, thin-walled, rarely branched, 3–5 μm diam.
Hymenium: Cystidia and cystidioles absent; basidia narrowly clavate, with four sterigmata and a basal clamp connection, 24–28 × 4–5.5 µm.
Basidiospores: Cylindrical, colorless, thin-walled, smooth, IKI–, CB–, (8.2–)8.5–10.3(–10.5) × 3–4 µm, L = 9.40 µm, W = 3.66 µm, Q = 2.51–2.63 (n = 60/2).
Type of rot: White rot.
Additional specimens examined: CHINA, Yunnan Province, Chuxiong, Zixishan Forestry Park, E 101°24′, N 25°0′, alt. 1950 m, on angiosperm stump, 26 Jun 2018, CLZhao 6731 (SWFC).
Hyphoderma mopanshanense C.L. Zhao, sp. nov. Figs. 5, 6.
MycoBank no.: MB 836262.


Holotype: China, Yunnan Province, Yuxi, Xinping, Mopanshan National Forestry Park, E 101°29′, N 23°56′, alt. 2200m, on fallen angiosperm trunk, 19 Jan 2018, CLZhao 6498 (SWFC).
Etymology: Mopanshanense (Lat.): referring to the locality (Mopanshan) of the type specimens.
Basidiomata: Annual, resupinate, ceraceous when fresh, turn to hard ceraceous upon drying, up to 7 cm long, 5 cm wide, 100–300 µm thick. Hymenial surface porulose to pilose, white to slightly pale gray when fresh, slight gray to cream upon drying. Sterile margin indistinct, white to slightly gray.
Hyphal structure: Hyphal system monomitic, generative hyphae bearing clamp connections, IKI–, CB–, tissues unchanged in KOH.
Subiculum: Generative hyphae colorless, thick-walled, rarely branched, 4–6 μm diam.
Hymenial layer: Generative hyphae colorless, thick-walled, rarely branched, 3.5–6.5 μm diam.
Hymenium: Cystidia fusiform, numerous, thick-walled, septated, apically encrusted with crystal, 86–171 × 10.5–13 µm, cystidioles absent; basidia clavate, with four sterigmata and a basal clamp connection, 15–18.5 × 3–4.5 µm.
Basidiospores: Cylindrical, colorless, thin-walled, smooth, IKI–, CB–, (7.6–)7.8–9.7(–10) × 2.6–3.3 µm, L = 8.78 µm, W = 2.95 µm, Q = 2.77–2.98 (n = 60/2).
Additional specimens examined: CHINA, Yunnan Province, Yuxi, Xinping County, Mopanshan National Forestry Park, E 101°29′, N 23°56′, alt. 2200m, on the fallen branch of angiosperm, 4 Jan 2018, CLZhao 6493 (SWFC).
3.2. Molecular phylogenyThe ITS dataset included sequences from 33 fungal specimens representing 19 taxa. The dataset had an aligned length of 647 characters in the dataset, of which 310 characters are constant, 47 are variable and parsimony-uninformative, and 290 are parsimony-informative. Maximum parsimony analysis yielded 1 equally parsimonious tree (TL = 1238, CI = 0.459, HI = 0.541, RI =0.646, RC = 0.296). Best model for ITS estimated and applied in the Bayesian analysis: GTR+I+G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis resulted in the similar topology with an average standard deviation of split frequencies =0.009955.
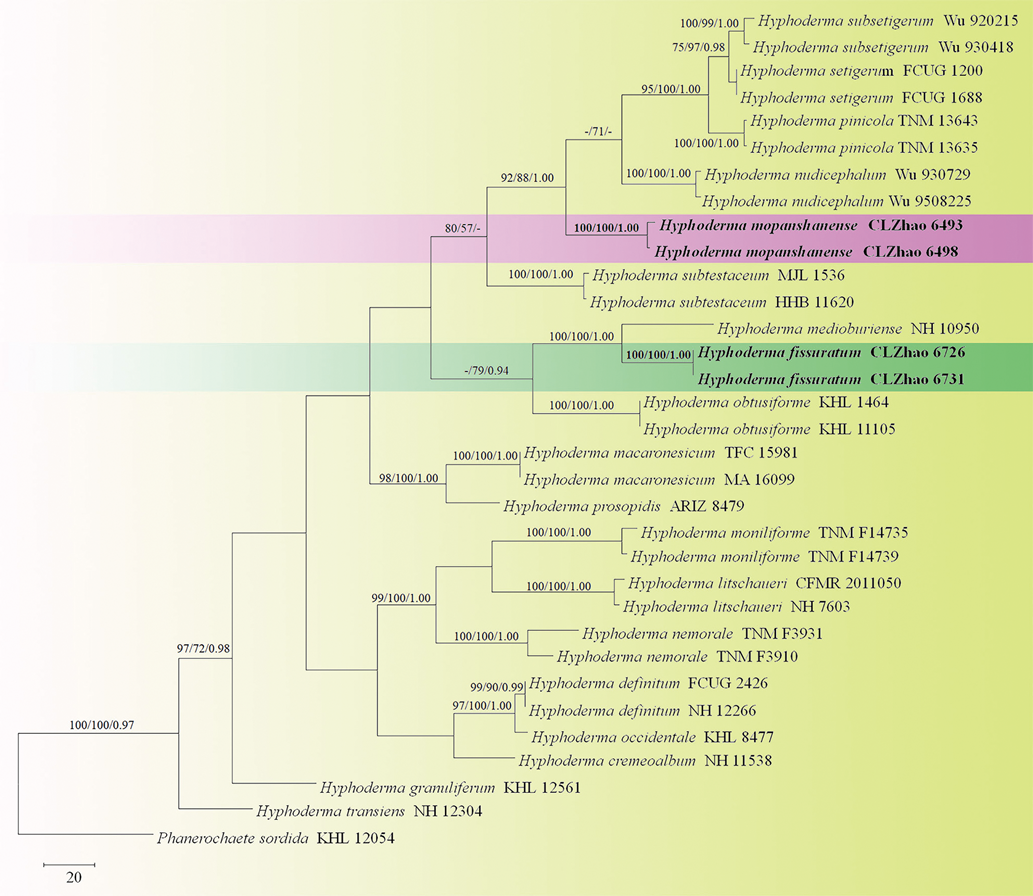
The phylogeny (Fig. 1) inferred from ITS sequences demonstrated that two new species, Hyphoderma fissuratum and H. mopanshanense, clustered into the Hyphoderma. Hyphoderma fissuratum was sister to H. medioburiense (Burt) Donk, and H. mopanshanense formed a monophyletic lineage with a strong support (100% BS, 100% BP and 1.00 BPP).
The nLSU dataset included sequences from 25 fungal specimens representing 19 species. The dataset had an aligned length of 1300 characters, of which 931 characters are constant, 168 are variable and parsimony-uninformative, and 201 are parsimony-informative. Maximum parsimony analysis yielded 4 equally parsimonious trees (TL = 792, CI = 0.605, HI = 0.395, RI = 0.491, RC = 0.297). Best model for the nLSU dataset estimated and applied in the Bayesian analysis: GTR+I+G, 1set nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis and ML analysis resulted in a similar topology as MP analysis, with an average standard deviation of split frequencies = 0.009036 (BI).
The phylogenetic tree (Fig. 2) inferred from nLSU sequences revealed that Hyphoderma fissuratum grouped with H. roseocremeum (Bres.) Donk, and H. mopanshanense was sister to H. setigerum.
In the present study, two new species Hyphoderma fissuratum and H. mopanshanense was described based on phylogenetic analyses and morphological characters.
Phylogenetically, Hyphoderma fissuratum was sister to H. medioburiense based on ITS sequence data (Fig. 1). However, morphologically, Hyphoderma medioburiense differs from H. fissuratum by its yellowish ochraceous hymenial surface, presence of cystidia and larger basidiospores (11–15 × 4–5 µm; Bernicchia & Gorjón, 2010). In the nLSU sequences, Hyphoderma fissuratum was grouped with H. roseocremeum, but morphologically H. roseocremeum differs in its whitish to yellowish ochraceous hymenial surface with rose tint and presence of tubular cystida (Bernicchia & Gorjón, 2010).
Morphologically, Hyphoderma fissuratum reminds on five species based on lacking cystidia, H. acystidiatum Sheng H. Wu, H. cremeoalbum (Höhn. & Litsch.) Jülich, H. densum Sheng H. Wu, H. sibiricum (Parmasto) J. Erikss. & Å. Strid and H. singularibasidium Dhingra, Avn.P. Singh & Singla. However, H. acystidiatum differs in its white hymenial surface and grandinioid hymenophore and wider basidiospores (9–11.5 × 4.5–5.3 µm; Wu, 1997); H. cremeoalbum in its presence of paraphysoid hyphae among the basidia and larger basidiospores (10–14 × 5–6 µm; Jülich, 1974); H. densum in its ivory yellow hymenial surface and larger basidiospores (10.5–12.5 × 4.2–5 µm; Wu, 1997); H. sibiricum in its strikingly yellow hymenial surface and wider basidiospores (7–9 × 4–5 µm; Eriksson & Ryvarden, 1975); H. singularibasidium in its grayish white to yellowish white hymenial surface and presence of the lateral outgrowth from the middle of basidium (Dhingra, Singh, & Singla, 2009).
In phylogenetic study, Hyphoderma mopanshanense formed a single lineage with good support (100% BS, 100% BP and 1.00 BPP) and then grouped with H. subtestaceum (Litsch.) Donk and a clade comprising taxa of H. nudicephalum Gilb. & M. Blackw., H. pinicola Yurch. & Sheng H. Wu, H. setigerum, H. subsetigerum Sheng H. Wu on the basis of ITS sequences (Fig. 1). In the nLSU sequences, H. mopanshanense has sister to H. setigerum and then clustered with H. nudicephalum and H. pinicola (Fig. 2). However, H. nudicephalum differs from H. mopanshanense by having the farinaceous to odontioid hymenial surface, conspicuous capitate cystidia with nonincrusted apices and wider basidiospores (7–9 × 3.5–4 µm; Gilbertson & Blackwell, 1988). Hyphoderma pinicola differs in its chalky white hymenial surface and larger basidiospores (13–16 × 4–4.5 µm; Yurchenko & Wu, 2014b). Hyphoderma setigerum is separated from H. mopanshanense by the smooth to tuberculate hymenophore, larger basidia (25–30 × 6–75 µm) and basidiospores (7–11 × 3–4.5 µm; Bernicchia & Gorjón, 2010). Hyphoderma subsetigerum differs in its grandinioid hymenophore with whitish to ivory yellow hymenial surface and smaller basidiospores (6–8 × 2.8–3.2 µm; Wu, 1997). Hyphoderma subtestaceum differs from H. mopanshanense by having the membranaceous hymenophore with pallidus to cream to isabellinus hymenial surface (Litschauer, 1928).
In the geographical distribution, both new species, Hyphoderma fissuratum and H. mopanshanense were collected at the same spot (Yunnan Province). However, they are not grouped together in ITS and nLSU analyses (Fig. 1, 2), and H. fissuratum differs from H. mopanshanense by having the cracking hymenial surface and larger basidiospores (8.5–10.3 × 3–4 µm).
Wood-rotting fungi is an extensively studied group of Basidiomycota (Núñez & Ryvarden, 2001; Bernicchia & Gorjón, 2010; Dai 2012; Ryvarden & Melo, 2014; Dai et al., 2015), but the Chinese wood-rotting fungus diversity is still not well known, especially in subtropics and tropics, many recently described taxa of this ecological group were from these areas (Dai, 2012; Chen, Korhonen, Li & Dai, 2014; Bian & Dai, 2015; Cui et al., 2019; Shen et al., 2019; Zhu, Song, Zhou, Si & Cui, 2019). Two new species in the present study are from subtropics, too. It is possible that a number of other new taxa will be found after further field studies and molecular analyses.
Disclosure
The authors declare no conflict of interest. All the experiments undertaken in this study comply with the current laws of the People's Republic of China.
The research is supported by the National Natural Science Foundation of China (Project No. 31700023) and the Science Foundation of Southwest Forestry University Projects (Nos. 111715, QN201904) and the Science Foundation of Department of Education of Yunnan Province Projects (Nos. 2020Y0396, 2020Y0395).