2021 Volume 62 Issue 4 Pages 239-243
2021 Volume 62 Issue 4 Pages 239-243
The obligate biotrophic oomycete genus Pustula is one of the four major linages of white blister rusts (Albuginaceae) identified so far. Species of the genus Pustula cause white blister rust on numerous genera in the asterids, represented by several phylogenetically distinct genus-specific lineages, most of which still await formal description. Thus, the observation of the species of Pustula on the Asteraceae subfamily Gymnorhenoideae pointed out to the existence of a hitherto undescribed species. By the morphological and molecular phylogenetic investigation conducted in this study it is concluded that the pathogen on Gymnarrhena micrantha from Iran indeed represents a hitherto unknown species and is described as P. persica. This species has apparently adapted to desert condition and is, after Albugo arenosa, the second species of white blister rust from Iranian deserts, highlighting the adaptability of white blister rusts to hot and dry habitats.
Despite similarities, such as filamentous growth and osmotrophic nutrient uptake, the phylum Oomycota is unrelated to fungi of the kingdom Mycota, but instead belongs to the kingdom Straminipila, which also contains diatoms and brown seaweeds (Beakes & Thines, 2017). Organisms in the Oomycota have adapted to a wide range of climate conditions and lifestyles (Thines, 2014) and can be found in both arctic habitats (Hassett, Thines, Buaya, Ploch, & Gradinger, 2019) and hot deserts (Mirzaee et al., 2013). The highest diversity has so far been found among the two independently-evolved, obligate biotrophic lineages parasitizing angiosperm plants, the downy mildews and white blister rusts (Thines, 2014; Wijayawardene et al., 2020). The white blister rusts have evolved to sporulate below the epidermis of their hosts and to liberate their spores by enzymatic digestion of the epidermal layer covering the pustules (Heller & Thines et al., 2009). The family Albuginaceae contains the three genera that cause white blister disease of angiosperms, Albugo, Pustula, and Wilsoniana. The latter two have been segregated from Albugo based on their largely different cytology, differences in sporangia and oospore morphology, as well as deep phylogenetic divide (Thines & Spring, 2005). In total, there are four major lineages in the Albuginaceae (Voglmayr & Riethmüller, 2006), each with a specific host range. Albugo s.str. parasitizes mostly Brassicales, but a few lineages are present on other orders (Choi, Shin, Ploch, & Thines, 2011b; Ploch, Choi, & Thines, 2018). Albugo s.l. parasitizes members of the Convolvulaceae and is distinguishable from Albugo s.str. by a pronounced oogonium ornamentation (Voglmayr & Riethmüller, 2006; Thines & Voglmayr, 2009). Wilsoniana is parasitic to caryophyllids and features broadly pear-shaped sporangia and densely ridged or reticulate oospores (Thines & Spring, 2005; Thines & Voglmayr, 2009). Pustula parasitizes various asterids, in particular Asteraceae, and is characterised by usually densely reticulate oospores and sporangia with an equatorial wall thickening (Thines & Spring, 2005; Choi, Thines, Tek, & Shin, 2012). Traditionally, it has been assumed that species causing white blister rust disease are specific mostly on the host family level (Wilson, 1907; Biga, 1955; Choi & Priest, 1995). However, phylogenetic investigations have revealed that in Albugo, besides the generalist species, A. candida (Pers.) Roussel, several distinct, host-specific species exist, which seem to be specific below the host genus level (Choi, Shin, Hong, & Thines, 2007; Choi, Shin, Ploch, & Thines, 2008; Choi, Shin, & Thines, 2009; Thines et al., 2009; Ploch et al., 2010; Choi & Thines, 2011). Also in the genus Pustula, species seem to be specific on at least the host genus level (Ploch et al., 2011), leading to the description of P. helianthicola C. Rost & Thines affecting sunflower (Rost & Thines, 2011) and the re-appraisal of several species previously thought to be synonyms of P. obtusata (Link) C. Rost (syn. P. tragopogonis (Pers.) Thines) (Choi et al., 2012) In line with this, three new species of Pustula were recently introduced from the Junggar Basin in China (Xu, Song, Xi, & Jiang, 2016; Xu et al., 2018).
During field trips in Iran, the occurrence of Pustula on Gymnarrhena micrantha Desf. was noticed. Gymnarrhena micrantha is a hardy member of Asteraceae growing in dry, mostly bare and sandy areas in the deserts of Iran. It is an ephemeric, amphicarpic, dwarf desert annual herb which is mainly distributed in the drier parts of the Mediterranean biome of North Africa and the Middle East. Although some variation across the distribution range has been noticed in collections, there is only one species recognized in the genus. In a study of the tribe Inuleae using the cpDNA gene ndhF, it was found that Gymnarrhena did not belonging to Asteroideae as previously thought, but rather to the paraphyletic Cichorioideae complex or sister to the entire Asteroideae,. Thus, it was, proposed as the sole member of the subfamily Gymnarrhenoideae (Anderberg, Eldenäs, Bayer, & Englund, 2005; Funk & Fragman-Sapir, 2009).
Given the host specificity previously observed for the genus Pustula (Ploch et al., 2011; Xu et al., 2016, 2018) it seemed plausible that the Pustula species occurring on Gymnarrhena does not belong to any Pustula species described so far. Therefore, it was the aim of this study to clarify the phylogenetic relationships of the potential new species and to investigate its morphology.
Specimens sequenced in this study have been deposited in the Herbarium Senckenbergianum in Frankfurt (international herbarium code FR). The collection details are given in Table 1. Thin cross sections using a razor blade were done on wetted herbarium specimens with white blister symptoms. Sections were transferred to 60% lactic acid or 5% aqueous chloral hydrate solution on a slide. The preparations for microscopy were warmed up, covered with coverslips and screened in bright-field using a compound light microscope (VWR TR 500 PH, VWR International, Darmstadt, Germany). Subsequently, suitable preparations were investigated in differential interference contrast (DIC) using a using a compound light microscope (Zeiss Imager2, Carl Zeiss, Jena, Germany) for measurements and photographs. Measurements were performed at ×1,000 magnification. Measurements are presented as (minimum–)mean minus standard deviation–mean–mean plus standard deviation(–maxiumum), with all values apart from the mean rounded to the nearest 0.5 µm increment, followed by the number of measurements done for the respective organ.
Pathogen |
Host |
Collection details |
GenBank accession # for cox2 (mtDNA) |
Herbarium accession # |
Pustula obtusata s.lat. |
Tragopogon graminifolius |
Iran, Birjand, Mohammadiah, leg. Mohammad Reza Mirzaee, Apr 2009 |
MW450686 |
FR0046065 |
P. obtusata s.lat. |
T. graminifolius |
Iran, Birjand, Mohammadiah, leg. Mohammad Reza Mirzaee, Apr 2009 |
MW450687 |
FR0046066 |
P. persica sp. nov. |
Gymnarrhena micrantha |
Iran, Ferdows, leg. Mohammad Reza Mirzaee, Apr 2010 |
MW450684 |
FR0046081 |
P. persica sp. nov. |
G. micrantha |
Iran, Khorsan, Mohammadiah, leg. Mohammad Reza Mirzaee, Apr 2009 |
MW450685 |
FR0046083 |
Genomic DNA was extracted from small pieces of leave tissue with pustules of Pustula from dried specimens. DNA extraction and PCR were performed as reported before (Mirzaee et al., 2013) In short, the innuPREP plant DNA extraction kit (Analytik Jena GmbH, Jena, Germany) was used for DNA extraction and PCR was performed using cox2 primers reported previously (Hudspeth, Nadler, & Hudspeth, 2000). PCR products were sequenced by the laboratory centre of the Senckenberg Biodiversity and Climate Research Centre, with the primers used in PCR. GenBank accession numbers for the sequences obtained in this study are given in Table 1.
The partial cox2 sequences from the specimens were edited using the DNASTAR computer package version 8 (Lasergene, Madison, WI, USA), and Geneious version 5.3.4 (Biomatters Ltd., Auckland, New Zealand). Subsequently, they were added to the dataset of Ploch et al. (2011). In addition, the sequences of two Pustula species recently described were added (Xu et al., 2016, 2018). Sequences were aligned on the Mafft webserver (Katoh, Rozewicki, & Yamada, 2019) using default settings. Phylogenetic analyses were done on the TrEase webserver (http://thines-lab.senckenberg.de/trease/) using FastTree2 (Price, Dehal, & Arkin, 2010) for Minimum Evolution inference, RAxML (Stamatakis, 2014) for Maximum Likelihood inference, both with 1,000 bootstrap replicates, and Bayesian inference using MrBayes, version 3.2 (Ronquist et al., 2012) with 5 Million generations, while other parameters were set to default.
In the phylogenetic reconstructions (Fig. 1), Pustula sp. from infecting Gymnarrhena micrantha is occupying an isolated position, with no clear affinities to any other lineage. The two specimens from G. micrantha were identical in sequence and clustered together with maximum support in all analyses. The specimens from P. obtusata s.lat. infecting Tragopogon graminifolius DC. clustered with P. obtusata from other species of Tragopogon with high to maximum support. However, some genetic divergence between the two groups was observed. Apart from a sister-group relationship of P. obtusata and P. junggarensis B. Xu & Z. D. Jiang, which received strong to maximum support, no other subdivisions in Pustula received strong support in all analyses.
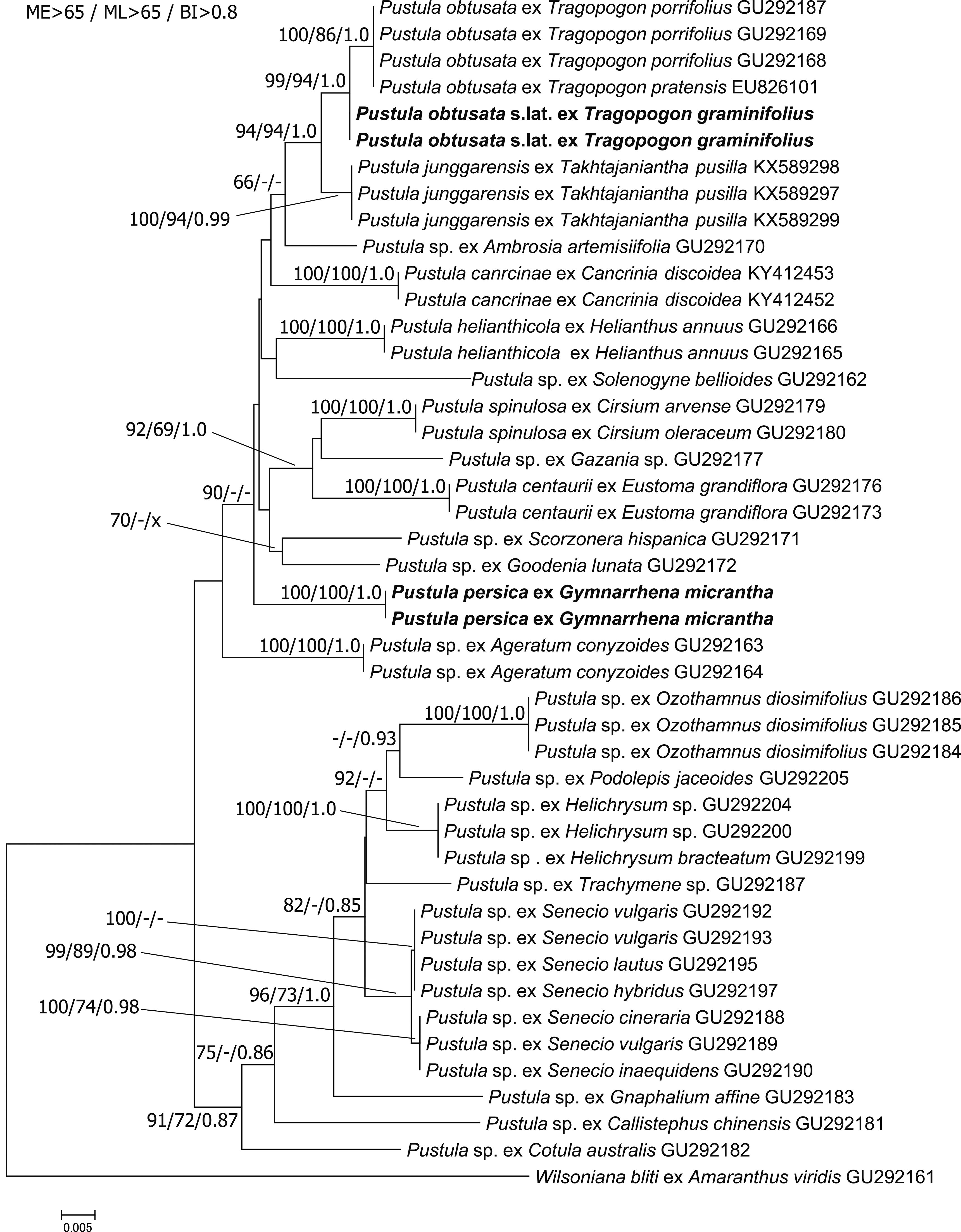
Pustula is a cosmopolitan genus of white blister rusts, reported from all continents except for Antarctica, and affecting a wide range of members of the Asterales ( Ploch et al., 2011). Most species of Pustula have been observed in the Asteraceae subfamilies Lactucoideae, Carduoideae and Asteroideae. So far, there has been no occurrence reported in the subfamily Gymnarrhenoideae (Funk & Fragman-Sapir, 2009), and also no other pathogen is known to affect genus Gymnarrhena. Therefore, Pustula persica, causing white blister rust disease, is the first pathogen reported in this subfamily. Interestingly, this is the forth species of Pustula described from arid regions (Xu et al., 2016, 2018), supporting that this genus is able colonise a broad range of climates and is well-adapted to dry conditions.
The morphological features that distinguish P. persica, introduced here, from other members of Pustula are a combination of oospore, oogonium, primary and secondary sporangia sizes, in line with previous publications (Rost & Thines, 2011; Choi et al., 2012; Xu et al., 2016, 2018). As especially the often-neglected primary sporangia seem to be informative (Constantinescu & Thines, 2006; Choi et al., 2012), these should always be considered when investigating species of Pustula.
While several new, host-specific species have been described or confirmed in the genus Albugo (Choi et al., 2007, 2008; Thines et al., 2009; Ploch et al., 2010, 2018; Choi & Thines, 2011; Choi, Thines, & Shin, 2011a), there are so far rather few articles on undescribed Pustula species (Ploch et al., 2011; Rost & Thines, 2011; Xu et al., 2016, 2018) Probably, this is because the variation in the oospore ornamentation is often less conspicuous than in Albugo (Ploch et al., 2010; Choi et al., 2012) As a result, with the exception of P. helianthicola (Rost & Thines, 2011), all of the undescribed lineages found inPloch et al. (2011) still await description. Apart from the species P. chardiniae (Bremer & Petr.) Thines, P. hydrocotyles (Petr.) Thines, P. obtusata (as P. tragopogonis), and P. spinulosa (de Bary) Thines, originally included in Pustula by Thines and Spring. (2005), only nine new species or combinations have been introduced to date, including P. obtusata, P. centaurii Thines, C. Rost & Y. J. Choi, P. helianthicola and P. obtusata (Rost & Thines, 2011), P. brasiliensis (Speg.) Y. J. Choi & Thines, and P. swertiae (Berl. & Kom.) Y. J. Choi & H. D. Shin (Choi et al., 2012), as well as P. junggarensis (Xu et al., 2016), P. cancriniae B. Xu & Z. D. Jiang, and P. xinyuanensis B. Xu, Y. J. Choi & Z. D. Jiang (Xu et al., 2018).
Generally, Pustula species seem to be host specific on the host genus level (Ploch et al., 2011; Choi et al., 2012), but in this study, we found some sequence divergence for P. obtusata s.lat. on T. graminifolius as compared to P. obtusata from other Tragopogon species. Further investigations, ideally including inoculation experiments and multigene phylogenies are necessary to elucidate, if this variation can be considered as an intra-species variation or if there are two distinct species of Pustula occurring in this host genus.
Taxonomy
Pustula persica Mirzaee & Thines, sp. nov. Fig. 2.
MycoBank no.: MB 838463.
Type: IRAN, Ferdows, on G. micrantha, Apr 2010, leg. M. R. Mirzaee (holotype, Herbarium Senckenbergianum FR0046081).
Gene sequence in GenBank: MW450684 (cox2).
Etymology: “persica” refers to the Latin name of Iran, the country in which the species was first collected.

Description: Hyphae intercellular. Sori mostly hypophyllous, 0.5–2 mm diam, distinct or confluent, rounded or irregular, whitish to cream in colour. Sporogenous hyphae hyaline, unbranched, clavate to cylindrical, head 13.5–16 μm diam, 25.5–39 μm long. Sporangia arranged in basipetal chains, hyaline, of two types. Primary sporangia hyaline with an overall thickened wall, except for the distal wall mostly 17–20.5 μm diam. Secondary sporangia hyaline, subglobose to cylindrical, with pronounced equatorial wall thickening, mostly 18–22 μm long, 18.5–23 μm diam. Oogonia globose to irregular, hyaline to yellowish, wall smooth, mostly 63–80 µm diam. Oospores globose, yellowish brown to dark brown, mostly 49.2–60.5 diam. Oospore ornamentation predominantly finely reticulate, sometimes appearing wrinkly and with a discontinuous, wider net and rounded protuberances with fine ridges, fine ornamentation with an irregularly bulged pattern within the areolae.
Habitat and known distribution: On living leaves of G. micrantha in Iran.
Other specimens examined: IRAN, Khorsan, Mohammadiah, April 2009, leg. M. R. Mirzaee, Herbarium Senckenbergianum FR0046083.
Notes: Sori of P. persica on G. micrantha were mostly hypophyllous, 0.5–2 mm diam, distinct or confluent, rounded or irregular, and whitish to cream in colour. The hyaline primary sporangia had an overall thickened wall, except for the distal wall, and measured (15–)17–20.5(–24) μm, 18.7 μm on average diam (n=100). The secondary sporangia were hyaline, subglobose to cylindrical, with a pronounced equatorial wall thickening. They were (15.5–)18–22(–27.5) μm,19.8 μm on average long (n=100) and (17–)18.5–23(–27.5) μm, 20.8 μm on average diam (n=90). Oogonia were globose to irregular, hyaline to yellowish, with a smooth wall, (57.7–)63–81(–89) μm, 72 μm on average diam (n = 41). Oospores were globose, yellowish brown to dark brown, (40.5–)49–60.5(–67.9) μm, 54.9 μm on average diam (n=70). Oospore ornamentation was predominantly finely reticulate, but sometimes also wrinkly and with a discontinuous wider net and protuberances with fine ridges. Apart from the generally finely reticulate coarse ornamentation, a fine ornamentation with an irregularly bulged pattern within the areolae was observed. A comparison with P. obtusata from T. graminifolius from Iran to P. persica on G. micrantha showed differences in oospore size. While the oospore size of P. obtusata was (42.5–)46–51.5(–56.5) μm, 48.7 μm on average diam, P. persica on G. micrantha, had larger oospores with (40.5–)49–60.5(–68) μm, 54.9 μm on average diam. Primary sporangia of P. obtusata were larger than in P. persica, with (15–)18–24.5(–30) μm, 21.2 μm on average diam in the former species and (15–)17–20.5(–24) μm, 18.7 μm on average diam in the latter.
The authors declare no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where they were performed.
Support by the Iranian Research Institute of Plant Protection (IRIPP) to MRM is gratefully acknowledged. This study was supported by the LOEWE initiative in the framework of the Biodiversity and Climate Research Centre (BiK-F). Lisa Nigrelli and Sabine Telle are gratefully acknowledged for laboratory support. Following the suggestion of Thines et al. (2020), all scientific names, regardless of rank, are given in italics. Author contributions – MRM and MT conceived the study, MRM provided materials, processed the specimens and did the microscopy work. SP and MRM did the laboratory work, MT performed phylogenetic reconstructions, MT and MRM wrote the manuscript with contributions from SP.