2020 年 37 巻 4 号 p. 123-132
2020 年 37 巻 4 号 p. 123-132
Although infection of Porphyromonas gingivalis (P. gingivalis) is closely correlated with progress of vascular inflammatory diseases, distinctive pathogenesis induced by P. gingivalis is still elusive. We investigated the impact of P. gingivalis lipopolysaccharide (P. gingivalis-LPS) on gene expression in endothelial cells with comparison to Escherichia coli lipopolysaccharide (E. coli-LPS) and zymosan. Here, we showed that the expressions of interleukin-6 and tumor necrosis factor-α mRNA induced by P. gingivalis-LPS are much lower than those by E. coli-LPS, and that the expressions of interleukin-8 and hypoxia inducible factor-2α are also lower than those by zymosan. These effects of P. gingivalis-LPS-mediated alterations of gene expression were cancelled in the absence of fetal bovine serum in culture medium. Of note, P. gingivalis-LPS treatment reduced the gene expressions of tissue factor and plasminogen activating inhibitor-1 as opposed to either E. coli-LPS or zymosan treatment. Our finding suggests that alterations of gene expression pattern in endothelial cells mediated by P. gingivalis-LPS may contribute to the pathogenesis of vascular inflammatory diseases.
It is well known that Porphyromonas gingivalis (P. gingivalis) is a major causative bacterium of periodontitis [ 1, 2]. Several studies have indicated that P. gingivalis implicates in the progression of several vascular inflammatory diseases such as atherosclerosis, sepsis, hypertension, vascular hemorrhage, ischemic stroke, non-alcoholic steatohepatitis, and Alzheimer’s disease (AD) [ 3– 7]. As the bleeding from the lesion with periodontitis has been constantly occurred, P. gingivalis is able to infiltrate from the periodontium to intravascular space and then disseminate systemic vascular vessels [ 8]. The recent animal model studies and clinical studies have demonstrated that P. gingivalis is detected in intra-vascular vessels and injured tissues, suggested that P. gingivalis plays a role in the pathogenesis of vascular inflammatory diseases [ 3, 8].
We previously reported that presence of P. gingivalis in the bloodstream and detachment of endothelial cells are important for the development of arteriosclerosis in vivo animal model [ 9]. We also clarified that the direct contact with P. gingivalis (and/or lipopolysaccharide) caused transformation of aortic smooth muscle cells and resulting in the acceleration of cell proliferation in vitro condition [ 10]. These findings suggest that normal condition of endothelial cells is necessary for prevention of P. gingivalis–induced vascular diseases such as arteriosclerosis [ 11, 12]. In contrast, direct effects of P. gingivalis to endothelial cell itself are still unclear.
Endothelial cells, embedded in inner layer of blood vessels, play crucial roles for the regulation of vascular function including vascular inflammation, blood coagulation, vascular permeability, vasodilation, and vasoconstriction. Thus, dysregulated activation of endothelial cells is an underlying cause of vascular inflammatory diseases, cardiovascular diseases, and neuroinflammatory diseases [ 13, 14]. Several studies indicated the possibility that intra-vascular infection of P. gingivalis potentially impairs endothelial functions leading to atherosclerosis that, associated with systemic inflammatory and hyper coagulant state [ 3, 5, 8, 13]. In addition, it has been thought that P. gingivalis dampens the endothelial cell barrier and endothelium structure and then enters subendothelial space [ 15]. Therefore, it is considered that P. gingivalis may cause the increase in vascular permeability. Of note, the spreading of P. gingivalis in intra-vascular/brain tissue is recognized as an important trigger of the onset of neuroinflammatory diseases [ 7, 16].
The lipopolysaccharide (LPS) of P. gingivalis is known as one of the major substances of this pathogen [ 17]. It has been reported that P. gingivalis-LPS induces pro-inflammatory responses in monocytes and periodontal fibroblasts [ 18, 19]. P. gingivalis-LPS contains O-antigen, core oligosaccharide, and lipid A as same as Escherichia coli (E. coli)-LPS, while the difference in the acylation patterns of lipid A from them has been identified [ 17]. Due to the structural and componential difference of LPSs, it is thought that P. gingivalis-LPS induced cell activation is distinct from E. coli-LPS [ 18, 20]. In agreement this, E. coli-LPS is well-known as toll like receptor (TLR)-4 agonist, whereas it has been reported that P. gingivalis-LPS acting as an agonist for TLR-2 or as an antagonist and/or agonist for TLR-4 activation [ 21- 23]. In addition, several studies indicated that P. gingivalis-LPS might induce differential inflammatory signaling dependent on cell type specific pattern [ 18, 24]. Thus, studies about the effect of P. gingivalis-LPS on endothelial cell hold promise for understanding how P. gingivalis triggers vascular inflammatory diseases.
In this study, we investigated the impact of P. gingivalis-LPS on the expression of pro-inflammatory and pro-coagulant genes in endothelial cells with comparison to TLR-4 agonist E. coli-LPS and TLR-2 agonist zymosan. We identified the differential expression pattern of pro-inflammatory and pro-coagulant genes in cultured endothelial cells induced by P. gingivalis-LPS.
Cell culture
A human hybrid endothelial cell line, EA.hy926 cell, was generated by the fusion of human umbilical vein endothelial cell with the human lung carcinoma cell line A549 cell [ 25]. EA.hy926 cells were kindly provided by Dr. Cora-Jean S. Edgell (University of North Carolina, Chapel Hill). Cells were cultured on collagen-coated tissue-culture dishes (Corning, San Jose, CA) in Dulbecco’s modified Eagle’s medium (DMEM) (Nacalai Tesque, Kyoto Japan) containing 10% fetal bovine serum (FBS), 50 units/ml penicillin (Nacalai Tesque), and 50 μg/ml streptomycin (Nacalai Tesque) and were maintained at 37°C in a 5% CO2 atmosphere.
Detection of TLR mRNA expression in EA.hy926 cell
EA.hy926 cells were seeded in 35 mm collagen-I coated dish (Corning, San Jose, CA) at 0.3 × 106 cells/dish and cultured until semi-confluent condition. Total RNA was extracted using Isogen II (Nippon Gene, Tokyo, Japan) from EA.hy926 cells following manufacturer’s instructions and then cDNA was prepared using the Super Script VILO cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). The cDNA was amplified by AmpliTaq Gold 360 DNA polymerase (Thermo Fisher Scientific). Polymerase chain reaction (PCR) conditions were as follows: 35 cycles of 96°C for 60 s, 58°C for 60 s, and 68°C for 60 s, using specific primers for human TLRs ( Table 1). The PCR products were separated on 1.0% (w/v) agarose gel by electrophoresis.
| Primers | Sequences | |
|---|---|---|
| TLR-1 |
Forward Reverse |
5’-CAGCGATGTGTTCGGTTTTCCG-3’ 5’-GATGGGCAAAGCATGTGGACCA-3’ |
| TLR-2 |
Forward Reverse |
5’-GATGGGCAAAGCATGTGGACCA-3’ 5’-ACACCAGTGCTGTCCTGTGACA-3’ |
| TLR-4 |
Forward Reverse |
5’- ACACCAGTGCTGTCCTGTGACA-3’ 5’-AGGTAGAGAGGTGGCTTAGGCT-3’ |
| TLR-6 |
Forward Reverse |
5’-ACTGACCTTCCTGGATGTGGCA-3’ 5’-TGACCTCATCTTCTGGCAGCTC-3’ |
EA.hy926 cell stimulation with P. gingivalis LPS, E. coli LPS and zymosan
In order to investigate the effect of P. gingivalis-LPS (SMB00610-1MG, Sigma-Aldrich, Saint Louis), E. coli-LPS (0111:B4, Sigma-Aldrich, Saint Louis) and zymosan (NBP2-26233, Novus Biologicals) on endothelial cells, we stimulated EA.hy926 cells with them with or without FBS condition. In present of FBS condition, cells were washed with PBS and then stimulated with 1 µg/mL and 10 µg/mL of P. gingivalis-LPS, E. coli-LPS, or zymosan at in DMEM containing 10% FBS for 4 hours. In absence of FBS condition, EA.hy926 cells were starved for overnight with FBS free DMEM media. Then, cells were stimulated with 1 µg/ml and 10 µg/mL of P. gingivalis-LPS, E. coli-LPS, or zymosan at in DMEM without FBS.
Determination of mRNA expression in EA.hy926 cell after stimulation
After stimulations, cDNA was prepared from each cell, the quantitative real time PCR (qRT-PCR) was performed by using Thunderbird SYBR qPCR Mix (Toyobo. Osaka, Japan). The relative quantitative gene expression was determined by following 2-ΔΔCt method and normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Specific primers for the detection of target genes were shown in Table 2.
| Primers | Sequences | |
|---|---|---|
| IL-6 |
Forward Reverse |
5’-ATGGGCTACAGGCTTGTCACTC-3’ 5’-TTCTGCCAGTGCCTCTTTGCTG-3’ |
| IL-8 |
Forward Reverse |
5’-GAGAGTGATTGAGAGTGGACCAC-3’ 5’-CACAACCCTCTGCACCCAGTTT-3’ |
| TNF-α |
Forward Reverse |
5’-CTCTTCTGCCTGCTGCACTTTG-3’ 5’-ATGGGCTACAGGCTTGTCACTC-3’ |
| HIF-2α |
Forward Reverse |
5’-CACAACCCTCTGCACCCAGTTT-3’ 5’-TTGCCATAGGCTGAGGACTCCT-3’ |
| TF |
Forward Reverse |
5’-CAGAGTTCACACCTTACCTGGAG-3’ 5’-GTTGTTCCTTCTGACTAAAGTCCG-3’ |
| PAI-1 |
Forward Reverse |
5’-CTCATCAGCCACTGGAAAGGCA-3’ 5’-GACTCGTGAAGTCAGCCTGAAAC-3’ |
| GAPDH |
Forward Reverse |
5’-GAAGGTGAAGGTCGGAGTC-3’ 5’-GAAGATGGTGATGGGATTTC-3’ |
Effect of P. gingivalis-LPS on permeability of endothelial cells
The permeability of endothelial cells assay was investigated with the slight modification of the previously established method [ 26]. First, the polycarbonate trans-well insert with a 0.3-μm (Corning, San Jose, CA) was coated with fibronectin and allowed for 30 minutes incubation at 37˚C. Then, EA.hy926 cells were seeded on trans-well inserts at a density of 2 × 105 cells with the DMEM medium. The cells were allowed to grow until completely the cells monolayer formed. The monolayer formation was confirmed by using phase contrast light microscopy.
In the presence of FBS, before stimulation with LPS, the cells were washed with 1x PBS for two times. Then, the lower chamber of unstimulated group was filled with 600 μL of 10% FBS DEMEM media with 200 μg/mL FITC dextran (Sigma-Aldrich, Saint Louis). The stimulation group’s lower chamber was filled with 600 μL 10% FBS media supplemented with 200 μg/mL FITC dextran and 10 μg/mL P. gingivalis LPS or E. coli LPS.
In FBS starvation condition, before stimulation with LPS, the cells were starved with DMEM medium without FBS for 12 hours after washing with 1x PBS for two times. Then, the lower chamber of control was filled with 600 μL of 0% FBS DEMEM media with 200 μg/mL FITC dextran. The lower chamber of stimulatory group was filled with 600 μL 0% FBS DMEM media containing 200 μg/mL FITC dextran and 10 μg/mL P. gingivalis LPS or E. coli LPS.
At indicated time points (30 min, 60 min, 120 min and 180 min), 20 μL aliquots of media from the upper chamber was withdrawn and diluted with 180 μL 1x PBS. Finally, after triplicating in black 96-well plates (Corning, San Jose, CA), the fluorescence intensity was measured with excitation 485 nm and emission 520 nm.
Statistical analysis
Results were expressed as the mean ± SEM. Statistical comparisons were performed using Dunnett’s t-test. The results were considered significantly different at P < 0.05.
Endothelial cell line EA.hy926 cell expresses TLR-4, TLR-2, TLR-1, and TLR-6
First, we confirmed the expression of TLR-4, TLR-2, and TLR2 co-receptors, TLR-1, and TLR-6, in EA.hy926 cells. EA.hy926 cells constitutively expressed TLR-4, TLR-2, TLR-1, and TLR-6 mRNA ( Fig. 1). In subsequent experiments, we used EA.hy926 cells and investigated the effect of P. gingivalis-LPS on several pro-inflammatory and pro-coagulant genes expression in endothelial cells.
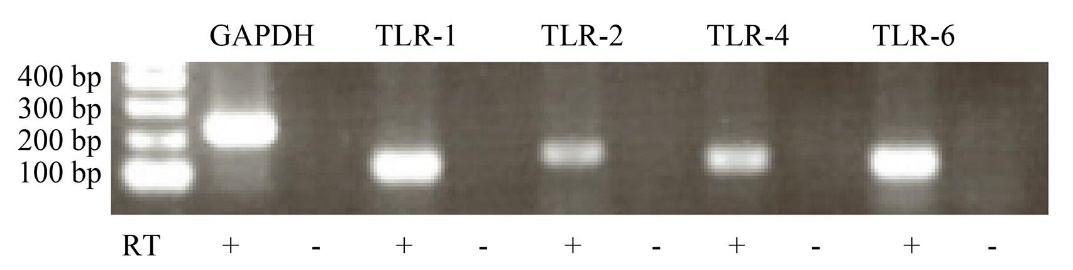
Total RNA was isolated from non-stimulated cells, and then cDNA was synthesized by RT reaction (RT+). The mRNA expression of TLR-1, TLR-2, TLR-4, and TLR-6 in EA.hy926 cells were detected by RT-PCR using specific primers as shown in table 1. GAPDH was used as an internal control. Representative image of three independent experiments is shown.
Effect of P. gingivalis-LPS on pro-inflammatory IL-6 and TNF-α mRNA expression
In order to investigate the impact of P. gingivalis-LPS on endothelial cells, we analyzed mRNA expression pattern in endothelial cells induced by P. gingivalis-LPS stimulation in comparison to TLR-4 agonist E. coli-LPS or TLR-2 agonist zymosan stimulation. We examined pro-inflammatory interleukin (IL)-6 and tumor necrosis factor-α (TNF-α) mRNA expressions that are typical genes induced by E. coli-LPS stimulation via TLR-4 activation in the presence of FBS. Although E. coli-LPS induced remarkably increases in IL-6 and TNF-α mRNA expressions, P. gingivalis-LPS induced weak expressions and zymosan did not ( Fig. 2).
To test whether serum components implicate in the effect of P. gingivalis-LPS, we examined IL-6 and TNF-α mRNA expression after treatment with P. gingivalis-LPS in the absence of FBS. Notably, both P. gingivalis-LPS-induced IL-6 and TNF-α mRNA expression in the absence of FBS were weaker than those in the presence of FBS, suggesting that serum components might contribute to the effect of P. gingivalis-LPS as like as E. coli-LPS ( Fig. 2).

EA.hy926 cells were stimulated with P. gingivalis-LPS, E. coli-LPS and zymosan in the presence (A) and absence (B) of FBS. The expressions of IL-6 and TNF-α mRNA in cells were evaluated by qRT-PCR. The results were normalized to GAPDH and data are presented as fold change comparing with unstimulated cells following 2-ΔΔCt method. Data were obtained from three independent experiments and are expressed as the mean ± SEM. *P < 0.05 and **P < 0.01 measured by Dunnett’s test compared to vehicle-treated cells.
Effect of P. gingivalis-LPS on IL-8 and HIF-2α mRNA expression
Next, we evaluated IL-8 and hypoxia inducible factor (HIF)-2α mRNA expression that induced by TLR-2 activation in the presence or absence of FBS. The stimulation of zymosan and E. coli-LPS significantly induced IL-8 expression in presence and absence of FBS ( Fig. 3). In contrast, P. gingivalis-LPS slightly induced IL-8 expression in the presence of FBS compared with zymosan and E. coli-LPS. The expression was not altered in the condition of the absence of FBS ( Fig. 3).
The expression of HIF-2α, a regulator of hypoxic adaptation involved in the inflammation, was upregulated by only zymosan stimulation in the presence of FBS, but not in the absence of FBS ( Fig. 3). Although P. gingivalis-LPS slightly induced HIF-2α in the presence of FBS, the effect of P. gingivalis-LPS was attenuated in the absence of FBS. The stimulation of E. coli-LPS did not induce the HIF-2α mRNA expression. These results indicated that P. gingivalis-LPS induces different patterns in IL-8 mRNA expression from zymosan and E. coli-LPS and represents similar expression pattern of HIF-2α mRNA by zymosan.
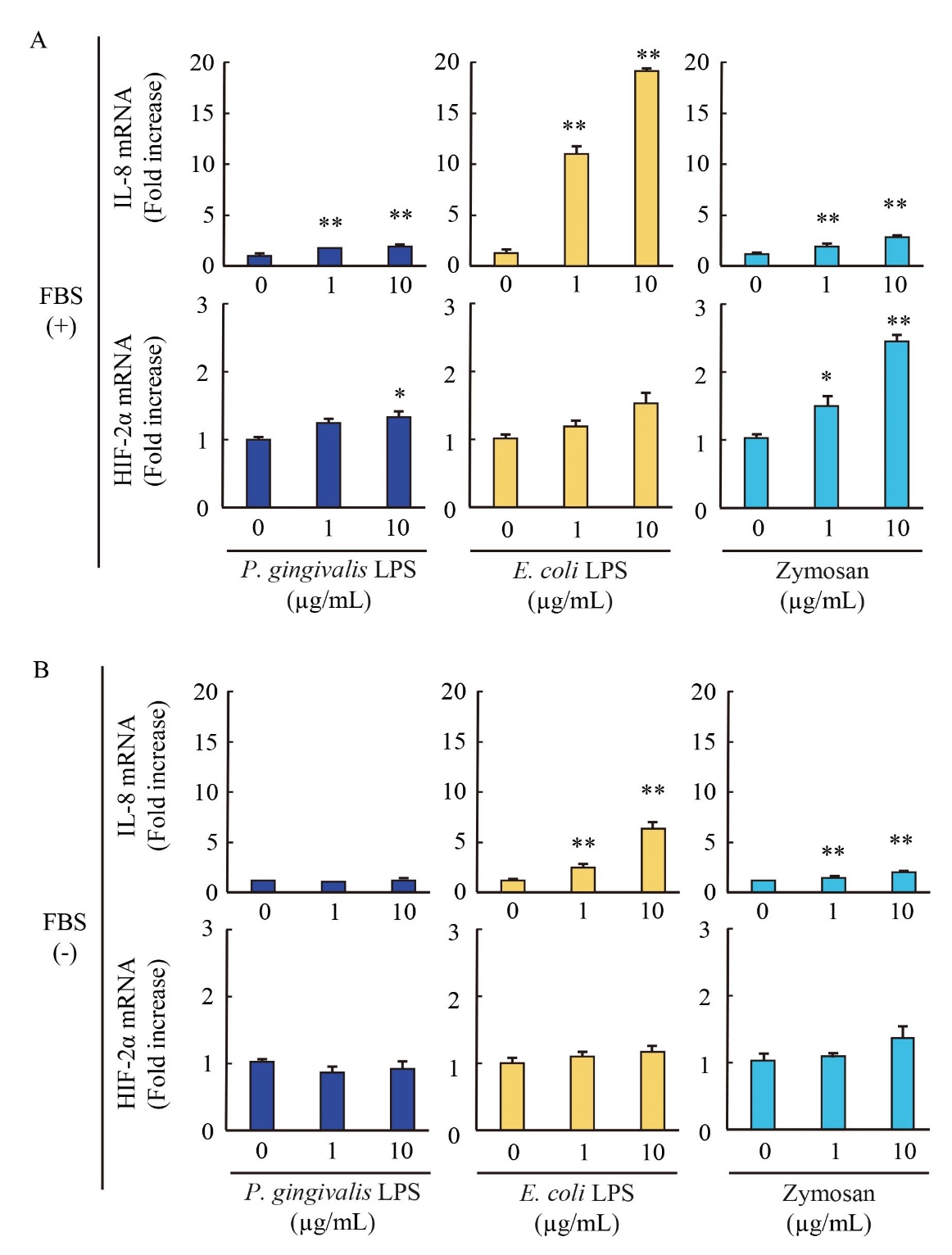
EA.hy926 cells were stimulated with P. gingivalis-LPS, E. coli-LPS and zymosan in the presence (A) and absence (B) of FBS. The expressions of IL-8 and HIF-2α mRNA in cells were evaluated by qRT-PCR. The results were normalized to GAPDH and data are presented as fold change comparing with unstimulated cells following 2-ΔΔCt method. Data were obtained from three independent experiments and are expressed as the mean ± SEM. *P < 0.05 and **P < 0.01 measured by Dunnett’s test compared to vehicle-treated cells.
Effect of P. gingivalis-LPS on pro-coagulant TF and PAI-1 mRNA expression
We explored the impact of P. gingivalis-LPS on mRNA expressions of pro-coagulant factors tissue factor (TF) and plasminogen activating inhibitor (PAI)-1 in endothelial cells. Notably, P. gingivalis-LPS significantly reduced both TF and PAI-1 mRNA expressions in the presence and absence of FBS ( Fig. 4). In our experimental condition, E. coli-LPS slightly induced those mRNA expressions in the absence of FBS, while zymosan did not alter the expressions with or without FBS. These results suggested that P. gingivalis-LPS represents anti-coagulant activity to endothelial cells.
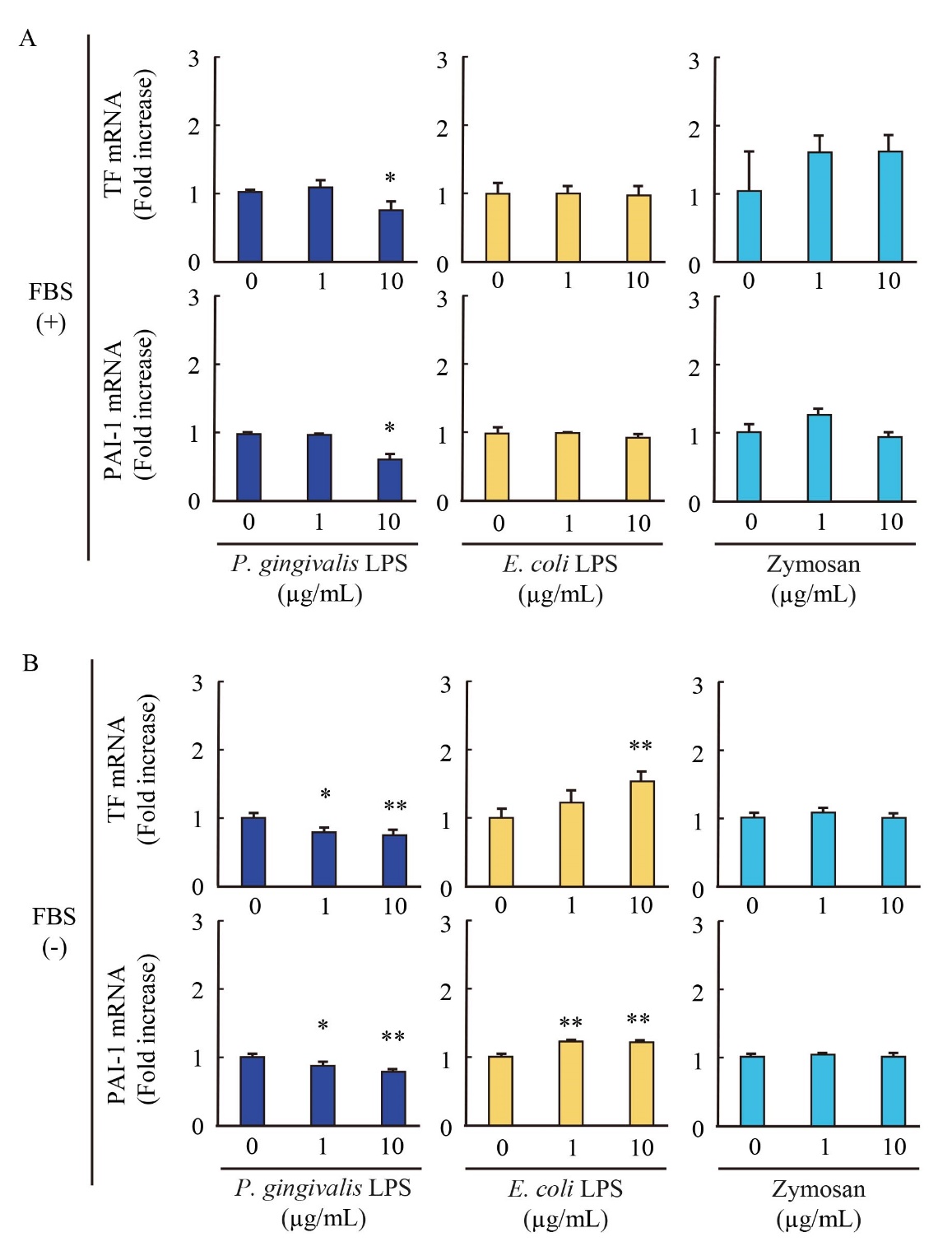
EA.hy926 cells were stimulated with P. gingivalis-LPS, E. coli-LPS and zymosan in the presence (A) and absence (B) of FBS. The expressions of TF and PAI-1 mRNA in cells were evaluated by qRT-PCR. The results were normalized to GAPDH and data are presented as fold change comparing with unstimulated cells following 2-ΔΔCt method. Data were obtained from three independent experiments and are expressed as the mean ± SEM. *P < 0.05 and **P < 0.01 measured by Dunnett’s test compared to vehicle-treated cells.
Effect of P. gingivalis-LPS on permeability of endothelial cells
Transwell cultured EA.hy926 cells were stimulated by P. gingivalis-LPS or E. coli-LPS in the presence or absence of FBS and measured the permeability of FITC-dextran. The endothelial cell permeability stimulated by P. gingivalis-LPS or E. coli-LPS was not altered in the presence and absence of FBS ( Fig. 5). These results suggested that bacterial LPS may not affect the endothelial cell permeability.

Transwell cultured EA.hy926 cells were stimulated by 10 µg/mL of P. gingivalis-LPS or E. coli-LPS with 200 μg/mL FITC dextran adding in the lower chamber in the presence (A) and absence (B) of FBS after confirming monolayer formation. At indicated time points (30 min, 60 min, 120 min, and 180 min) 20 μL aliquots of media was withdrawn from the upper chamber and diluted in 180 μL 1x PBS. After triplicating in black 96-well plate, the fluorescence intensity was measured with excitation 485 nm and emission 520 nm. Data were obtained from three independent experiments and expressed as the mean ± SD.
P. gingivalis is one of the pathogenic periodontopathic bacteria and is frequently detected in the periodontal pockets of patients with chronic periodontal disease [ 1]. Several studies have been indicated that chronic periodontitis implicated in the progression of cardiovascular diseases through the activation of systemic inflammation [ 27, 28]. The pathogens derived from P. gingivalis are known as inducers of pro-inflammatory cytokines expression such as IL-1β, IL-6, TNF-α, and matrix metalloproteinase-1 in human gingival fibroblast, monocytes, and macrophage cells [ 20, 29, 30]. These cells predominantly maintain homeostasis of gingival tissues by controlling inflammation, tissue repair, and regeneration, while also contribute to the stability and progression of systemic inflammation via the circulating system [ 31, 32]. Of note, P. gingivalis has properties of adhering to blood vessels and inducing inflammation, and those properties are directly involved in the vascular inflammation and promotion of cardiovascular diseases [ 33]. Therefore, analysis of the specific interaction of P. gingivalis with endothelial cells is required for understanding the pathogenesis of P. gingivalis in vascular diseases.
In this study, our results demonstrated that P. gingivalis-LPS induces differential expression of pro-inflammatory and pro-coagulant genes in endothelial cells in comparison to E. coli-LPS and zymosan. Such differential expression pattern may be due to the difference in degree of TLR-2 and TLR-4 activation based on their receptor complex formation. P. gingivalis-LPS is one of the major pathogens of P. gingivalis and is recognized as ligands for TLR-2 and TLR-4 in monocytes, macrophages, human gingival fibroblast and periodontal ligament stem cells [ 18, 20, 30, 34]. TLR-2 forms a heterodimer complex with the TLR-1 or TLR-6 and recognizes bacterial pathogens following their initial interaction with CD14 [ 35, 36]. TLR-4, is constitutively expressed in immune cell and endothelial cells, forms a complex with the myeloid differentiation-2 protein after the initial binding of bacterial LPS to CD14 [ 37]. These studies have reported that serum cofactors, CD14 and LPS-binding protein, contribute to TLR-2 or TLR-4 activation [ 38]. E. coli-LPS is a well-known ligand for TLR-4 and a critical inducer of septic shock and infectious systemic inflammation that is associated with endothelial cell dysfunction [ 39]. Meanwhile, TLR-2 is an important receptor detecting a large spectrum of microbial pathogen associated molecular patterns [ 35]. This feature of TLR-2 is characterized by receptor heterodimers with TLR-1 and TLR-6, which not only expands the range of pathogen associated molecular patterns but may also dictate the balance between tolerant and defensive responses [ 35]. Taken together, the constructional difference in TLRs complex in endothelial cells might be essential for P. gingivalis-LPS mediated gene expression.
It is reported that LPS of P. gingivalis is structurally different from that of E. coli [ 17]. The lipid A of P. gingivalis-LPS are mainly consisted by two acylated lipid A including tetra-acylation and penta-acylation, whereas, E. coli-LPS contains only hexa-acylated lipid A [ 17]. Due to this structural difference, it has been thought that P. gingivalis-LPS induces distinguished pathology via activation of either TLR-2 or TLR-4, and both resulting to cardiovascular diseases [ 40]. In this study, P. gingivalis-LPS induced pro-inflammatory cytokines IL-6 and TNF-α in endothelial cells in the presence of FBS as like as E. coli-LPS, although TLR-2 ligand zymosan did not induce them. Moreover, P. gingivalis-LPS slightly induced IL-8 and HIF-2α gene expressions that are target of TLR-2 signaling as same as zymosan. Additionally, E. coli-LPS remarkably induced IL-8 gene expression than zymosan and P. gingivalis LPS, while did not alter HIF-2α expression. Based on the present findings, we speculated that P. gingivalis-LPS partially activates both TLR-2 and TLR-4 on endothelial cells ( Fig. 6). Moreover, cofactors such as soluble CD14, CD36, and Dectin-1 are identified for ligand recognition of TLR-2 and TLR-4 [ 41]. These cofactors might sustain the interaction between TLRs and their ligand leading to the initiation of the signaling pathway. Our finding is that P. gingivalis LPS attenuates IL-6, TNF-α, IL-8, and HIF-2α mRNA expression in the absence of FBS suggested that serum cofactors are required for P. gingivalis-LPS-mediated endothelial cell activation ( Fig. 6). However, further studies will be needed to identify any cofactors involved in P. gingivalis-LPS-mediated endothelial cell activation.
In the current study, our data demonstrated that P. gingivalis-LPS reduces pro-coagulant tissue factor and PAI-1 mRNA expression in endothelial cells. It is thought that expression level of tissue factor and PAI-1 in endothelial cells directly links to intravascular blood coagulation state. Thus, our data suggest the possibility that P. gingivalis might modulate the balance of blood coagulation and fibrinolysis that involved in the development of neuroinflammation [ 42]. Yu et al. reported that plasmin is capable to cleave, degrade, and reduce both non‐aggregated monomeric and aggregated fibrillar amyloid-β forms [ 43]. Furthermore, plasmin protected neurons from amyloid-β‐induced cell death and enhanced the clearance of amyloid-β in AD animal models upon pharmacologically PAI‐1 inhibition [ 43]. Indeed, it has been shown that plasmin activity in AD human brain homogenates is reduced as compared to that of normal subjects [ 44]. Although P. gingivalis-LPS reduces PAI-1 mRNA expression in endothelial cells, since plasmin generation is determined in consequence of the balance of blood coagulation and fibrinolysis activity, P. gingivalis-LPS might provide a favorable environment for AD progression and neuroinflammation via attenuating pro-coagulant state.
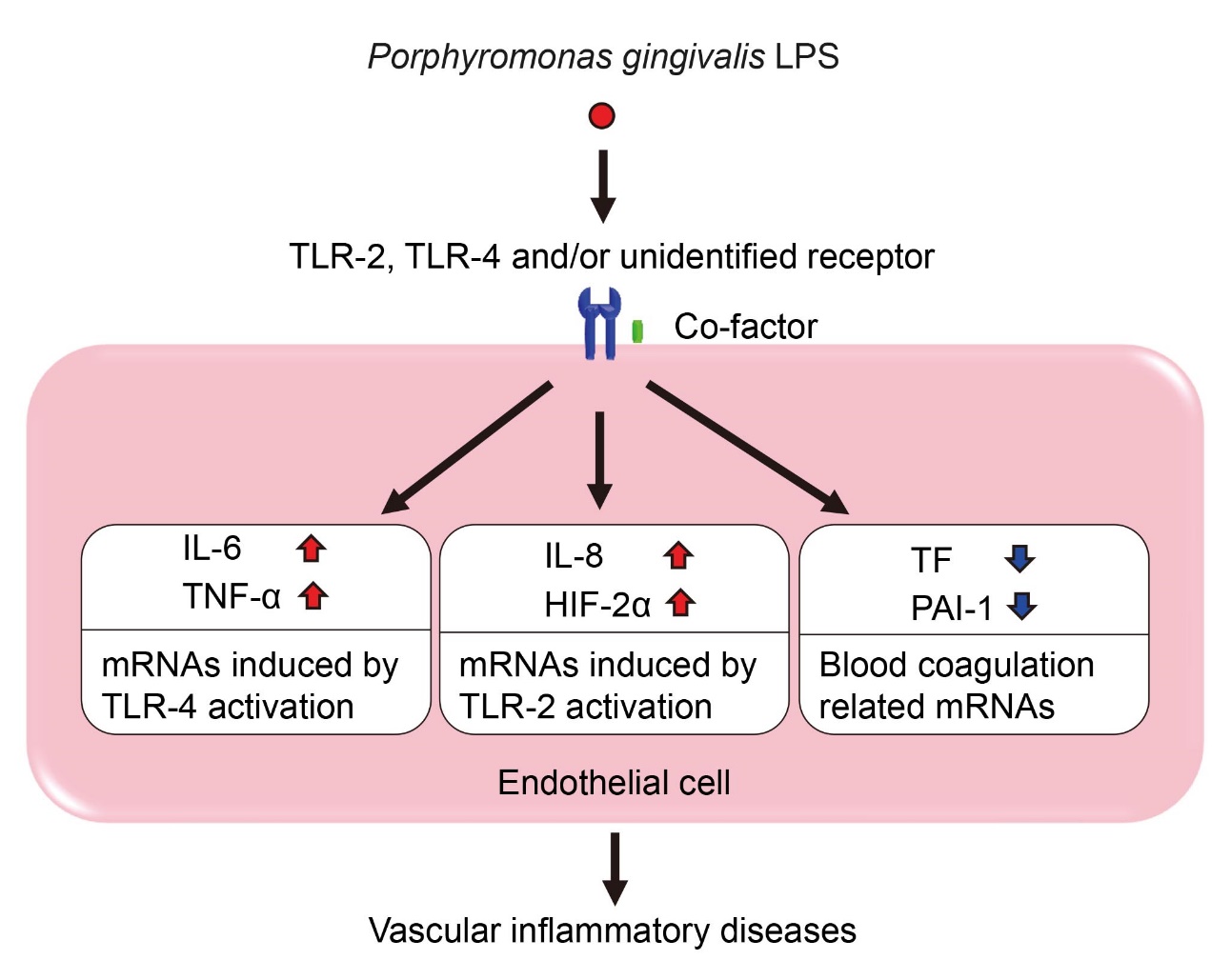
In this study, we investigated the differential gene expression pattern of P. gingivalis-LPS stimulation compared with stimulation of E. coli-LPS or zymosan. The differential gene expression pattern induced by P. gingivalis-LPS stimulation which is likely mediated by the activation of TLR-2 and TLR-4 may contribute to the development of the vascular inflammatory diseases.
Acknowledgements
We thank Prof. Atsushi Nagai, Shoma Kanda and Sonoko Karino for helpful discussions. This work was supported by JSPS KAKENHI (Grants-in-Aid for Scientific Research) [grant numbers, 19H04032, 19K10463, 19K08583, and 20K11509].
Ccnflicts of interest
The authors declare no conflicts of interest.