2014 年 37 巻 6 号 p. 1021-1028
2014 年 37 巻 6 号 p. 1021-1028
Tribulus terrestris fruits are well known for their usage in pharmaceutical preparations and food supplements. The methanol extract of T. terrestris fruits showed potent inhibition against the papain-like protease (PLpro), an essential proteolylic enzyme for protection to pathogenic virus and bacteria. Subsequent bioactivity-guided fractionation of this extract led to six cinnamic amides (1–6) and ferulic acid (7). Compound 6 emerged as new compound possessing the very rare carbinolamide motif. These compounds (1–7) were evaluated for severe acute respiratory syndrome coronavirus (SARS-CoV) PLpro inhibitory activity to identify their potencies and kinetic behavior. Compounds (1–6) displayed significant inhibitory activity with IC50 values in the range 15.8–70.1 µM. The new cinnamic amide 6 was found to be most potent inhibitor with an IC50 of 15.8 µM. In kinetic studies, all inhibitors exhibited mixed type inhibition. Furthermore, the most active PLpro inhibitors (1–6) were proven to be present in the native fruits in high quantities by HPLC chromatogram and liquid chromatography with diode array detection and electrospray ionization mass spectrometry (LC-DAD-ESI/MS).
Specific proteolytic enzymes are key regulators of a wealth of physiological processes in humans. Similarly, pathogenic viruses, parasites and bacteria that cause infectious disease also rely upon proteolytic processing to complete their own life cycles and to serve as virulence factors.1) Most notably, two cysteine protease, a papain-like protease (PLpro) and 3C-like protease (3CLpro) play a critical role in virus-mediated RNA replication.2) Severe acute respiratory syndrome coronavirus (SARS-CoV) is a representative RNA virus that causes a highly contagious respiratory disease.3,4) The genome of SARS-CoV is translated into two large replicase polyproteins called as pp1a and pp1ab.5,6) PLpro and 3C-like protease domains present within these polyproteins direct their processing into 16 non-structural proteins (nsp1–16) that assemble to generate a multifunctional, membrane-associated replicase complex.7) The SARS-CoV PLpro performs the essential function of cleaving junctions spanning nsp1 through nsp4. Besides viral peptide cleavage, recent structural and functional studies demonstrated that PLpro is involved in a number of other important biochemical events, such as deubiquitination deISGylation, and evasion of the virus from the innate immune response.8–10) Thus, PLpro so serves as a model for development of drugs against other deubiquitinating enzymes involved in human diseases.11) Importantly, endogenous deubiquitinating enzymes are now strongly linked to cancer and neurological disease development and in spite of a huge research effort, no deubiquitinating enzyme inhibitors are as yet even in clinical trials.12)
Tribulus terrestris Linn belongs to the family Zygophyllaceae and is distributed in warm regions throughout India and the southern part of China. It is renowned as a saponin-rich plant with other known components being flavonoids, alkaloids, lignanamides and cinnamic amides.13–16) Given the wealth of interesting compounds contained within it, T. terrestris has been used in traditional Chinese medicine for the treatment of many diseases including sexual dysfunction.17,18) For instance, there is a substantial amount of research linking application of extracts of this plant to improvement in male and female libido disorders, relieving impotence and increasing sperm motility.17) The fruits, flowers and leaves are a very potent diuretic and tonic. It is also reported that saponins exhibit cytotoxic and antihyperlipidemic effects.19,20) Many pharmaceutical preparations and food supplements based on the saponin fraction of this plant are on sale worldwide.17)
Many synthetic amide derivatives have been prepared as potential PLpro inhibitors, but no natural amide compounds have yet been shown to exhibit PLpro inhibition.10,11) This encouraged us to search for PLpro inhibitors from T. terrestris because although it is most well known as a source of saponins, it is also known to contain cinnamic amides.15) In this study, we isolated six SARS-CoV PLpro inhibitors from the fruits of T. terrestris using bioactivity guided fractionation. The structures of these isolated compounds were identified as cinnamic amides using spectroscopic methods. The isolated amides were evaluated separately for their inhibitory activities against PLpro. Their inhibition mechanisms were ascertained using Lineweaver–Burk and Dixon plots. We also assessed the relative abundance of these extracts within the fruits using an HPLC chromatogram profile.
Organic solvents used for isolation were of first grade and the stock solution and buffers were prepared with milli Q water. Analytical grade methanol, acetonitrile and acetic acid for HPLC were purchased from J. T. Baker (Phillipsburg, NJ, U.S.A.). Column chromatography was carried out using silica gel (230–400 mesh, Merck), RP-18 (ODS-A, 12 nm, S-150 µM, YMC), and Sephadex LH-20 (Amersham Biosciences). Fluorescence spectra were measured on a SpectraMax M3Multi-Mode Microplate Reader (Sunnyvale, CA, U.S.A.). 1H- and 13C-NMR, as well as two dimensional (2D) NMR data, were obtained on a Bruker AM 500 (1H-NMR at 500 MHz, 13C-NMR at 125 MHz) spectrometer (Bruker, Karlsruhe, Germany) in CD3OD or DMSO-d6 with tetramethylsilane (TMS) as internal standard. Electron ionization-mass spectra (EI-MS), high resolution (HR)-EI-MS and FAB-MS, HR-FAB-MS were obtained on a JEOL JMS-700 mass spectrometer (JEOL, Tokyo, Japan). Qualitative analyses were made using an Agilent 1100 liquid chromatography (Agilent Technologies, Palo Alto, CA, U.S.A.). LC/MS was measured in a 3200 Q Trap LC/MS/MS System (Applied Biosystems, Lincoln, U.S.A.). Reagent grade chemicals were purchased from Sigma Chemical Co. (St. Louis, U.S.A.). The fruits of T. terrestris [imported from China, as permitted by Korea Food and Drug Administration (KFDA)] were purchased from a local market.
Preparation of ExtractThe fruits of T. terrestris were extracted in separate flasks (2 g dry fruits each) with 20 mL of ethyl acetate, methanol, 50% methanol in water and distilled water at room temperature for 1 week to examine the enzymatic inhibitory activities against SARS-CoV PLpro as a function of solvent used (Supplemental information). The methanol extract gave the strongest inhibition and it was used for further studies.
HPLC-ESI-MS/MS AnalysisQuantification of relative abundance of compounds assayed in this manuscript within the crude fruits extract was carried out by HPLC (Agilent 1100 series, Agilent Technologies) using a Zorbax Bonus-RP column (4.6×150 mm, 5 µm, Agilent Technologies). Absorbance was measured at 320 nm, About 10 µL of 10 mg/mL of crude fruits were loaded onto the column. The mobile phase for HPLC consisted of solvent A, 0.1% acetic acid in water, and solvent B, acetonitrile. The solvent gradient was as follows (relative to solvent A): 0 min, 0% B; 5 min 10% B; 12 min 20% B; 27 min 25% B; 38 min 30% B; 50 min 35% B.
MS/MS experiments were performed using a 3200 Q TRAP LC/MS/MS system (Applied Biosystems, Forser, CA, U.S.A.) with a Turbo VTM source and a Turbo ion Spray probe (500°C). The mass spectrometer was operated in positive and negative ion mode. Nitrogen was used as a nebulising and as well as a drying gas. The flow rates in both cases were 45 psi. The capillary voltage was set at 5.5 kV and the source temperature at 500°C. The resolutions of the first and third quadrupole were between 0.6 and 0.8 (unit resolution). Mass spectra were recorded between mass-to-charge ratio (m/z) 100 and 1000 with a step size of 0.1 amu.
Extraction and IsolationThe dried fruits (2.5 kg) of T. terrestris were macerated and extracted (10 L×3) for one week in total at room temperature, and then filtered. The filtrate was concentrated in vacuum to yield a dark green gum (312 g, 12.5%). The methanol extract (312 g) was washed with n-hexane (3 L×3, sonication) to remove oils to give subextract (118 g). A part of the extract (10 g) was fractionated (3 times) on reversed-phase (RP) silica gel column (340 g, C18) using MPLC (Puriflash 450, Interchim, Montlucon, France) with a linear gradient of 0–90% CH3OH/H2O and a 40 mL/min flow rate to afford seven fractions (A–G). Fractions C (1.3 g), E (2.8 g) and F (1.2 g) were grouped together and fractionated via MPLC using a silica gel column (200 g) with elution using a gradient of increasing CH3OH (0–15%) in CHCl3 to give subfractions A–H. Subfraction B (190 mg), enriched with 3 was further chromatographed over a C18 column (15 g) to afford compound 3 (38 mg). Subfraction C (460 mg), enriched with 2 and 4 was further chromatographed over a C18 column (30 g) to afford compounds 2 (47 mg) and 4 (32 mg). Subfractions D–E (625 mg), enriched with 1 and 6, were further chromatographed over Sephadex LH-20 (CH3OH) to give subfractions E1–E7. Subsequent separation of subfraction E3 (130 mg) by RP-MPLC using CH3OH−H2O (40 : 60) as mobile phase afforded compound 1 (25 mg). Subfraction E4 (108 mg) was chromatographed over a C18 column (15 g) using CH3OH−H2O (50 : 50) to give compound 6 (15 mg). Subfraction F (225 mg) was purified using Sephadex LH-20 column chromatography, eluting with CH3OH to afford compounds 5 (26 mg) and 7 (12 mg).
Expression and Purification of SARS-CoV PLpro from Escherichia coliThe PLpro gene (945 bp) was polymerase chain reaction (PCR) amplified from the plasmid pSARS-REP and cloned into a pPRoEx HT expression vector (Invitrogen, Carlsbad, CA, U.S.A.) which was subsequently transformed into DH5α competent cells. The expression plasmid was constructed such that PLpro carried an N-terminal His6-tag followed by a Tobacco etch Virus protease (TEV) cleavage site. Correct clones containing the PLpro gene in a pPRoEX HT vector were identified and verified by PCR and restriction digestion with BamHI and XhoI. The plasmid pPRoEx HT harbouring the PLpro gene was transformed into BL21 (DE3) E. coli (Novagen, Madison, WI, U.S.A.). Protein expression and purification was carried out according to a previous report.21) The protein purity was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and native-PAGE. The protein concentration was determined by Bradford assay using bovine serum albumin as the standard.22) The N-terminal his-tag was removed by TEV digestion prior to activity assays.
SARS-CoV PLpro Inhibition AssayIC50 values for all inhibitors were determined using a 96-well plate-based assay similar to our previously reported procedures. The substrate used in the assay was the fluorogeneic peptide Z-Arg-Leu-Arg-Gly-Gly-AMC (Z-RLRGG-AMC), which was purchased from ENZO Life Sciences. The substrate contains the five C-terminal residues of human ubiquitin with a C-terminal 7-amido-4-methylcoumarin (AMC) group. Hydrolysis of the AMC-peptide bond dramatically increases the fluorescence of the AMC moiety, allowing conversion to be accurately determined. Reactions were performed in a total volume of 200 µL, which contained the following components: 20 mM Tris-buffer, pH 8.0, 4 mM dithiothreitol (DTT), 30 µM Z-RLRGG-AMC, 2% dimethyl sulfoxide (DMSO), and varying concentrations of inhibitor (0–200 µM). Assays were initiated with the addition of PLpro to produce a final enzyme concentration of 60 nM. Reaction progress was monitored continuously on a SpectraMax M3Multi-Mode Microplate Reader (λexcitation=360 nm; λemission=460 nm; gain=40) and the reaction rate was calculated by the equation vi=vo/(1+[I]/IC50) using the enzyme kinetics module of Sigma Plot (v. 9.01 Systat Software, Inc.) where vi is the reaction rate in the presence of inhibitor, vo is the reaction rate in the absence of inhibitor, and [I] is the inhibitor concentration.
Protease Inhibition Mechanism StudiesThe kinetic study of SARS-PLpro used RLRGG-AMC as the substrate. The data acquisition was initiated within 1 min, and the reaction was monitored continuously every 30 s for up to 10 min. To study the kinetics of PLpro inhibition by compounds 1–6, various concentrations of compounds 1–6 were added to PLpro in assay buffer containing the predetermined substrate. Like the other kinetic trials, the data acquisition was initiated within 1 min, and the reaction was monitored continuously every 30 s for up to 10 min. All reactions were run in at least triplicate, and the results are presented as means±S.D.
Statistical AnalysisAll measurements were made in at least triplicate. The results were subject to variance analysis using Sigma plot. Differences were considered significant at p<0.05.
The extracts were obtained using four different polar solvent as described in Materials and Methods. These extracts were tested for enzyme inhibitory activity against SARS-CoV PLpro. To assess the inhibitory potency of each fraction against PLpro, activity was assayed according to a standard literature procedure. The SARS-CoV PLpro (residues 154−1855) was expressed in E. coli and purified by successive nickel affinity, ion-exchange and gel filtration chromatography (Supplemental information). The last step was used to separate the active monomer from larger oligomers. The mass of the isolated PLpro was confirmed by matrix assisted laser desorption/ionization-time-of-flight (MALDI-TOF). The His6-tag used for purification was removed by TEV cleavage. Activity of the purified PLpro was demonstrated by its ability to cleave a fluorogenic peptide substrate. The apparent Michaelis constant Km (27±0.9 µM) that is concentration of substrate at 1/2 Vmax was determined by plotting the initial rates corrected for the enzyme concentration (60 nM) versus substrate concentration (5–320 µM), and fitting the data to the Michaelis model (Supplemental information). All extracts investigated, apart from the water extract, showed inhibitory activity against PLpro at 300 µg/mL. In particular, methanol was found to be the solvent which gave maximum extraction of PLpro inhibitory substances. The composition of phytochemicals in each of the extracts was analyzed by HPLC with absorbance measured at 320 nm. A greater number of phytochemicals was detected in the HPLC chromatogram of the methanol extract than the others in this study (Supplemental information). Therefore, we chose methanol to extract PLpro inhibitors from fruits of T. terrestris.
Structural Identification of PLpro InhibitorsWe conducted phytochemical investigations to isolate bioactive compounds from the MeOH extract of the fruits of the target plant. This was achieved via repeated column chromatography over silica gel, octadecyl-functionalized silica gel and Sephadex LH-20. These efforts led to the isolation of seven compounds. The structure identification of these compounds was carried out by comparison with published data15,23) and spectroscopic analyses (Supplemental information). Compounds 1–7 were identified as N-trans-caffeoyltyramine (1), N-trans-coumaroyltyramine (2), N-trans-feruloyltyramine (3), terrestriamide (4), N-trans-feruloyloctopamine (5) and ferulic acid (7) including new compound (6) which we called terrestrimine (Fig. 1).
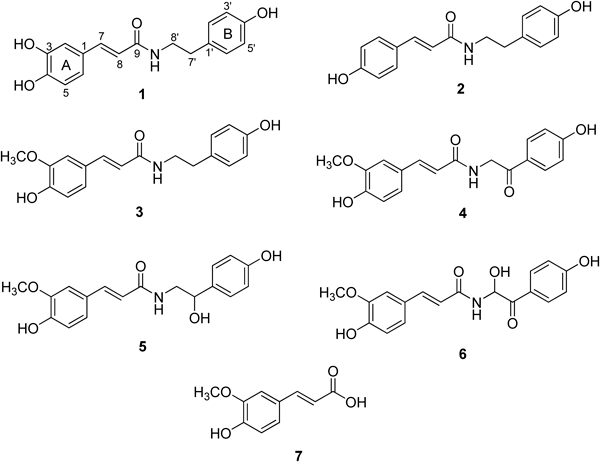
The structural elucidation of new compound 6 is detailed as follows (Table 1). Compound 6 was obtained as a yellow powder having the molecular formula C18H17NO6 and 11° of unsaturation established by HR-FAB-MS (m/z 344.1143 [M+H]+, calcd for C18H18NO6 344.1134). The compound 6, which has a chiral centre at C-8′, may exist as a racemic mixture because its [α] was almost zero. 1H- and 13C-NMR data in conjunction with distortionless enchancement by polarization transfer (DEPT) experiments indicated the presence of 18 carbon atoms, consisting of following functional groups: 1 methine (sp3), 9 methines (sp2), 1 methyl and 7 quaternary carbons. The 13C-NMR data enabled carbons corresponding to 7 C–C double bonds and 2 carbonyl groups to be identified, thus accounting for 9 of 11 the degrees of unsaturation. The extra two degrees of unsaturation were ascribed to two aromatic rings. The presence of a cinnamate group was easily deduced from strong connectivity between H-7 (δH 7.55, J=15.6 Hz) and H-8 (δH 6.51, J=15.6 Hz) in the correlation spectroscopy (COSY) spectral data and heteronuclear multiple bond connective spectroscopy (HMBC) correlation of H-8 with C-1 (δC 128.5), C-7 (δC 144.1) and C-9 (δC 169.1, carbonyl). HMBC correlation of the peak at δH 3.89 (OCH3) with C-3 (δC 149.8) unveiled the location of the methoxy moiety. Additionally the position of C4-OH (δC 150.7) was deduced from HMBC correlations of C4-OH with H-2 (δH 7.16) and H-6 (δH 7.07). The trisubstituted aromatic moiety of A-ring was affirmed by ortho coupling between H-5 (δH 6.82, J=8.2 Hz) and H-6 (δH 7.07, J=8.2 Hz), and meta coupling of H-2 (δH 7.16, J=1.6 Hz). The para-hydroxy phenyl moiety within the B-ring was easily affirmed by COSY correlation of H-2′/6′ (δH 7.98, 2H) and H-3′/5′ (δH 6.88, 2H), as well as ortho coupling (J=8.8 Hz) of both protons and HMBC correlation of H-3′/5′ with a quaternary carbon at C-4′ (δC 165.1). The carbinol moiety was confirmed by lower chemical shifts of C-8′ (δC 73.6) and H-8′ (δH 6.58) that agree with those typically observed for carbinolamines. HMBC correlation of H-8′ with C-7′ (δC 194.9) and C-1′ (δC 126.9) confirmed side chain of amide to be a 1-hydroxy-2-(4-hydroxyphenyl)-2-oxoethyl group. Strong HMBC correlation of H-8′ and C-9 (δC 169.1) proved the connection of the side chain within the cinnamate function (Fig. 2A). Thus, using the above obtained spectral data compound 6 was identified as (E)-N-(1-hydroxy-2-(4-hydroxyphenyl)-2-oxoethyl)-3-(4-hydroxy-3-methoxypheny) acrylamide which we called as terrestrimine. Moreover, the EI-MS of compound 6 showed fragment ions expected of a carbinolamine. The fragment ion at 325 [M−H2O]+ was prominent, indicated that water can be readily lost under strong ionizing conditions of the mass spectrometer. Other prominent peaks at m/z 193 [M−150]+ and 121 [M−222]+ can be ascribed to fragmentation by a McLafferty rearrangement as depicted in Fig. 2B. Importantly, decisive fragment (side chain) was observed at m/z 149 [M−H−193]− in electrospray ionization (ESI)-MS using the negative mode.
| Position | δH (mult, J in Hz) | δC (ppm) | HMBC |
|---|---|---|---|
| 1 | 128.5 | ||
| 2 | 7.16 (1H, d, J=1.6 Hz) | 112.2 | C-4, 6, 7 |
| 3 | 149.8 | ||
| 4 | 150.7 | ||
| 5 | 6.82 (1H, d, J=8.2 Hz) | 116.9 | C-1, 3, 4 |
| 6 | 7.07 (1H, dd, J=8.2, 1.7 Hz) | 124.1 | C-2, 4, 7 |
| 7 | 7.55 (1H, d, J=15.6 Hz) | 144.1 | C-1, 2, 6, 8, 9 |
| 8 | 6.51 (1H, d, J=15.6 Hz) | 118.6 | C-1, 7, 9 |
| 9 | 169.1 | ||
| 1′ | 126.9 | ||
| 2′, 6′ | 7.98 (2H, d, J=8.8 Hz) | 133.2 | C-3′, 4′, 5′, 7′ |
| 3′, 5′ | 6.88 (2H, d, J=8.8 Hz) | 116.9 | C-1′, 2′, 4′, 6′ |
| 4′ | 165.1 | ||
| 7′ | 194.9 | ||
| 8′ | 6.58 (1H, s) | 73.6 | C-1′, 7′, 9 |
| 3-OCH3 | 3.89 (3H, s) | 56.9 | C-3 |
All isolated cinnamamides (1−6) inhibited PLpro in a dose-dependent manner with IC50s ranging between 15.8 and 70.1 µM (Table 2, Fig. 3A). However, ferulic acid was inactive against PLpro up to 200 µM. New compound, terrestrimine (6) was found to be the most potent inhibitor with an IC50 of 15.8 µM. The potency of these inhibitors was affected by subtle changes in structure. It appears that better inhibition was observed when at least one of the methylene groups (C8′ and C7′) bore a polar substituent (ketone or alcohol). This is apparent from a comparison of compound 3 (IC50=70.1 µM) and others 4−6 (IC50=15.8−26.6 µM). The hydroxyl group on C-3 in ring A was not pivotal for PLpro inhibition as compound 1 and 2 had similar activities.
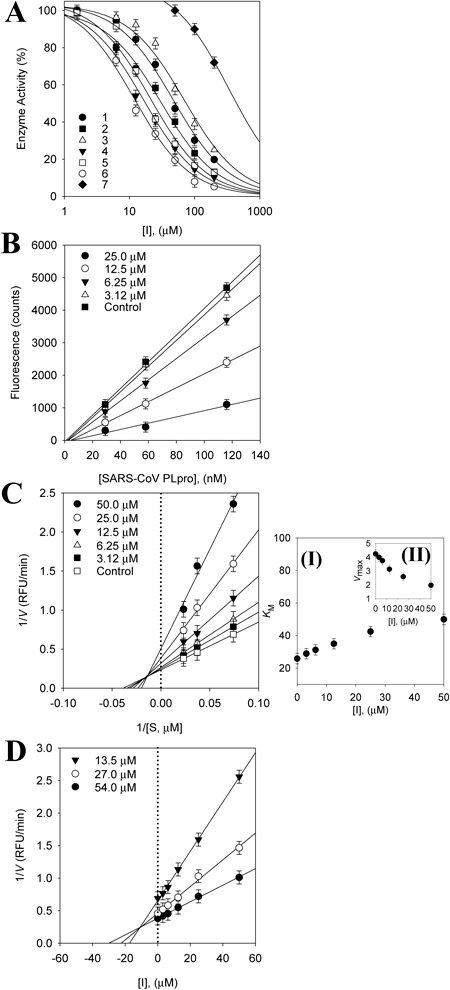
(B) The catalytic activity of SARS-CoV PLpro as a function of enzyme concentration at different concentrations of compound 6. (C) Lineweaver–Burk plot for the inhibition of compound 6 on the hydrolytic activity of PLpro. (D) Dixon plots of SARS-CoV PLpro inhibition by compound 6. Inset (I) Km values as a function of the concentrations of 6. (II) Dependence of Vmax on the concentration of 6.
Kinetic assays were repeated in the presence of different concentrations (1–200 µM) of compounds (1–6) to characterize inhibition of Z-RLRGG-AMC hydrolysis. All inhibitors manifested a similar relationship between enzyme activity and concentration. The inhibition of PLpro by compound 6 (the most effective species) is illustrated in Fig. 3A, representatively. Plots of the initial velocity versus enzyme concentration in the presence of different concentrations of compound 6 gave a family of straight lines, all of which passed through the origin. Increasing the inhibitor concentration resulted in lowering of the slope of the line, indicating that compound 6 is a reversible inhibitor (Fig. 3B). Other tested inhibitors (1–5) showed a similar profile to 6. The equilibrium constant for inhibitor binding, dissociation constant by inhibition (Ki) was obtained from the value at the intersection of three lines from Dixon plots (Fig. 3D). The Ki values of all compounds fell within the range 10.1–36.7 µM (Table 2). The enzyme inhibition properties of these derivatives were modeled using double-reciprocal plots. As depicted in Fig. 3C, the inhibition kinetics analyzed by Lineweaver–Burk plots show that compound 6 is a mixed-type inhibitor because increasing inhibitor concentration resulted in a family of lines which intersected at a non-zero point on both the x-axis and y-axis. Lineweaver–Burk plot showed that all compounds display mixed type inhibition because Vmax decreased, while Km increased with increasing concentrations of inhibitors (Supplemental information). All other inhibitors showed a similar kinetic mode.
| Compound | SARS-CoV PLpro | |
|---|---|---|
| IC50a) (µM) | Kinetic mode (Ki,b)µM) | |
| 1 | 44.4±0.6 | Mixed (24.5±0.6) |
| 2 | 38.8±0.4 | Mixed (20.1±1.3) |
| 3 | 70.1±0.7 | Mixed (36.7±1.1) |
| 4 | 21.5±0.5 | Mixed (13.9±0.8) |
| 5 | 26.6±0.5 | Mixed (16.2±1.0) |
| 6 | 15.8±0.6 | Mixed (10.1±0.5) |
| 7 | 200 < | NTc) |
a) All compounds were examined in a set of experiments repeated three times; IC50 values of compounds represent the concentration that caused 50% enzyme activity loss. b) Values of inhibition constant. c) NT is not tested.
To set the importance of these inhibitors in a practical content, we performed a comparative analysis of the levels of individual PLpro inhibitory cinnamamides within the native fruit using an LC-DAD chromatogram. The nature of each peak in the chromatogram was doubly verified by comparison with retention time of the pure compound (Supplemental information) and also by LC-ESI/MS analysis. The chromatography profile of peaks (1–7) showed a maximal absorption at 320 nm. Moreover, most PLpro inhibitors 1, 2, 3, 4 and 5 were present in very high concentrations because they appeared as the principal peaks in the chromatogram. The each peak was characterized from in the positive or negative ion modes by using liquid chromatography with diode array detection and electrospray ionization mass spectrometry (LC-DAD-ESI/MS) (Table 3 and Fig. 4). The MS spectrum of peaks 2 (tR=30.31, [M+H]+ at m/z 300.3), 4 (tR=37.68, [M+H]+ at m/z 314.2) and 6 (tR=39.75, [M+H]+ at m/z 284.3) were identical to our assigned structure for cinnamic amides (1–3), respectively. The major fragmentation of [M+H]+−137 in compounds 1–3 showed the loss of a 2-(4-hydroxyl phenyl)ethyl amine. Subsequently, the peaks 1 (tR=22.95, [M−H]− at m/z 328.2), 3 (tR=31.27, [M−H]− at m/z 342.2) and 5 (tR=38.15, [M+H]+ at m/z 328.4) were for cinnamic amides (4–6), respectively.
| Peak number | UV λmax (nm) | tR (min) at 320 nm | Molecular ion [M±H]+/− (m/z) | Fragment ions in ESI/MS (m/z) | HR-MS | Identification |
|---|---|---|---|---|---|---|
| 1 | 318 | 22.95 | 328.2 [M−H]− | 175.1 [M−H]−C8H9O2N1 | 329.1263 | N-trans-Feruloyloctopamine (5) |
| 2 | 318, 284 | 30.31 | 300.3 [M+H]+ | 163.1 [M+H]+C8H10O1N1 | 299.1158 | N-trans-Caffeoyltryamine (1) |
| 3 | 320, 291 | 31.27 | 342.2 [M−H]− | 177.1 [M−H]−C8H7O3N1 | 343.1134 | Terrestrimine (6) |
| 4 | 317, 283 | 37.68 | 314.2 [M+H]+ | 177.1 [M+H]+C8H9O2N1 | 313.1314 | N-trans-Feruloyltryamine (3) |
| 5 | 318, 286 | 38.15 | 328.4 [M+H]+ | 177.2 [M+H]+C8H8O2N1 | 327.1107 | Terrestriamide (4) |
| 6 | 312, 289 | 39.75 | 284.3 [M+H]+ | 147.1 [M+H]+C8H10O1N1 | 283.1208 | N-trans-Coumaroyltyramine (2) |
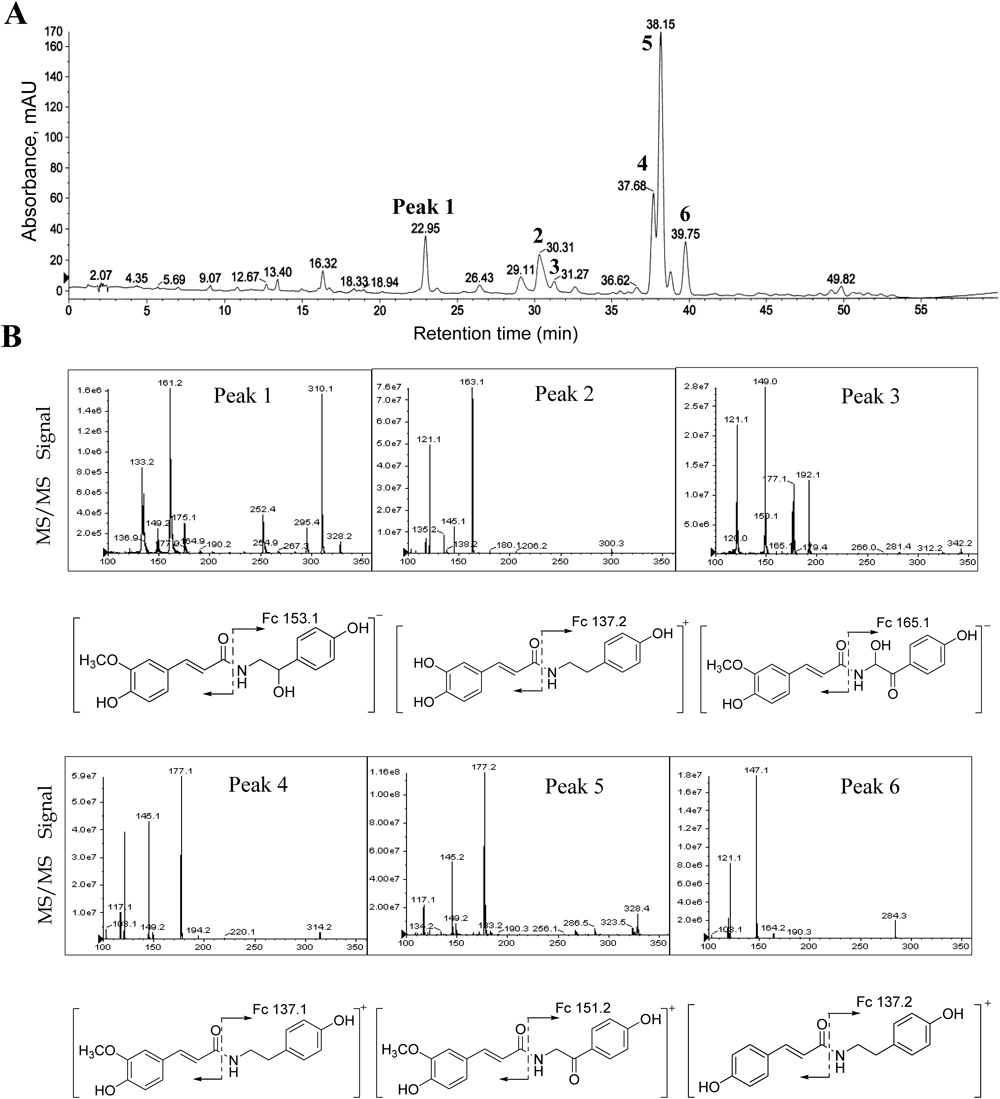
Peaks 1, N-trans-feruloyloctopamine; 2, N-trans-caffeoyltyramine; 3, terrestrimine; 4, N-trans-feruloyltyamine; 5, terrestriamide; 6, N-trans-coumaroyltyramine.
This study demonstrates that the methanol extract of T. terrestris fruits shows potent inhibitory activity toward SARS-CoV PLpro. Purification of this fraction gave six cinnamic amides including a novel cinnamic amide (6), which has an unusual carbinolamide motif. The inhibitory potencies and capacities of these amides toward papain-like protease (PLpro) were studied in detail. All compounds displayed significant inhibitory activity with IC50s values of 15.8–70.1 µM. All compounds showed mixed type inhibitory behavior. The most active PLpro inhibitors (1–6) were proven to be present in the native fruits in high quantities by HPLC and LC-DAD-ESI/MS. Given the huge importance of PLpro inhibitors in food and medicinal chemistry, we believe that these lead compounds could be of great significance to research in this pervasive field.
This research was supported by National Research Foundation Grant founded by Korea government (MEST) (2013M3A9A6003180) and a Grant from the Next-Generation BioGreen 21 Program (SSAC, NO. PJ009571), Rural Development Administration, Republic of Korea. MJCL acknowledges an HHMI international student research fellowship. All students were supported by a scholarship from the BK21 plus program.