2016 年 39 巻 2 号 p. 207-214
2016 年 39 巻 2 号 p. 207-214
Heart failure represents a major health problem. The development of new drugs to treat this condition is essential. We previously discovered that AF-001 attenuates the cardiac defects caused by heart failure in zebrafish. In this paper, we report the identification of AF-HF001, an AF-001 derivative, and its effects on live cardiomyocytes subjected to oxidative damage. The in vitro results demonstrated that AF-HF001 attenuates the production of reactive oxygen species (ROS) and the myocardial cell apoptosis. A DNA microarray was performed to broadly analyze gene expression after H2O2 treatment with or without AF-HF001. Hierarchical clustering analysis revealed that AF-HF001 modifies the expression of certain genes (Ndufs2, Ndufb6, Ndufb8, Ndufa13, Ndufs3, Ndufs5, TPM1, MYH14, RyR1, and TIMP4) related to ROS production, cardiac contractility and extracellular matrix remodeling. AF-HF001 ameliorates oxidative damage, which may be related to the mitogen-activated protein kinase (MAPK) family and the intrinsic mitochondrial pathway. Altogether, this study suggests that AF-HF001 exhibits potential as a clinical drug candidate for the treatment of heart failure.
Heart failure is a complex clinical syndrome with high prevalence, high in-hospital rates, significant healthcare costs and high mortality. Thus, developing new drugs for the treatment of heart failure will be beneficial. Unfortunately, the advances in drug discovery for heart failure have been minimal in recent years. Our research team discovered a positive inotropic compound, AF-001, in a phenotype-based, zebrafish model of heart failure; AF-001 decreased the heart and venous congestion size and effectively increased the blood flow velocity in caudal artery during in zebrafish after 2 h of treatment.1) We further discovered a derivative of AF-001, which is referred to as AF-HF001 (the chemical structure is presented in Fig. 1).
During heart failure progression, the adaptation of structure and function occurs in a step-wise fashion. The cellular compensation ultimately fails over time. Some studies reported that changes in the biological properties of myocardial cells and the progressive change of these cells cause heart failure at the cellular level. Previous research has demonstrated that patients with end-stage heart failure exhibited apoptosis leading to cardiomyocyte loss, which may contribute to progressive myocardial dysfunction.2,3) However, previous studies have also demonstrated that the production of reactive oxygen species (ROS) and the level of oxidative stress increased both in patients and animals with heart failure. ROS (including free radicals, such as superoxide, hydroxyl radical and nitric oxide) promote cardiac remodeling after myocardial damage, leading to phenotypic changes in myocardial cells (hypertrophy and death), inducing abnormal regulation of calcium signaling during contraction, and changing the extracellular matrix and metabolism of cardiac fibroblasts cells, resulting in cardiac structural and functional abnormalities. A connection between ROS and heart failure has been hypothesized. Apparently, myocardial ischemia and/or reperfusion with increased ROS production cause progressive cell death. It is conceptually plausible that treating ROS-induced cardiac apoptosis may provide an innovative therapeutic approach to heart failure. A better understanding of the mechanisms by which ROS activate signaling pathways could lead to identify a helpful tool in identifying new drug targets for the treatment of heart failure in the future.
To determine the role of AF-HF001 in ROS-induced cardiac apoptosis, we used hydrogen peroxide (H2O2) and cobalt chloride (CoCl2) induced H9c2 rat cardiomyoblast apoptosis. The study used DNA chip technology and aimed to determine gene expression profiling in H9c2 cells during oxidative stress injury with or without AF-HF001. Comparing these gene expression profiles would be helpful for understanding the mechanisms by which ROS induce heart failure and AF-HF001 ameliorates heart failure.
H9c2 rat cardiomyocyte is subclone cell line derived from embryonic BD1X rat heart tissue. 6×106 H9c2 cells were seeded into 60 mm culture plates (Corning, NY, U.S.A.). H9c2 cells (Drum Tower Hospital of Nanjing University Medical School, Nanjing, China) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L glucose (HyClone, Thermo Scientific, South Logan, UT, U.S.A.) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Thermo Scientific), 100 U/mL penicillin and 100 µg/mL streptomycin (Gibco BRL/Invitrogen, Carlsbad, CA, U.S.A.) in a humidified atmosphere at 37°C in 5% CO2. Following incubation for 24 h, the cell density reached 90%. H9c2 cells were treated with the indicated concentrations of H2O2 (100 µM; Sigma, St. Louis, MO, U.S.A.) or CoCl2 (200 µM; Sigma) as stimulus for the desired time with or without pretreatment with AF-HF001 for an additional 12 h.
The Measurement of Intracellular ROSThe generation of intracellular ROS in H9c2 cells was measured using the green fluorescence probe 6-carboxy-2′,7′-dichlorofluorescein diacetate (DCF-DA; Sigma). The cells were placed in 35-mm cell culture slides (Corning) and treated with H2O2 for 30 min. Then, the slides were washed twice with phosphate-buffered saline (PBS) and stained with 10 µM DCF-DA at 37°C for 30 min. The fluorescence images were obtained using Leica automated microscopes (Leica DM2500-3HF-FL, Solms, Germany). Scale bars indicate 50 µm.
The Assessment of Apoptosis and Caspase 3 AssayCell death was determined after H9c2 cells were treated with H2O2 or CoCl2 for 24 h as described above. Apoptosis was assessed using a cell death detection enzyme-linked immunosorbent assay (ELISA) kit (Roche, Germany) for the qualitative and quantitative determination of the cytoplasmic histone-associated DNA fragments. Cleaved caspase 3 activity was measured by Western blot with an anti-caspase 3 antibody (1 : 750; Cell Signal Technology, Danvers, U.S.A.).
Western Blotting AnalysisH9c2 cells were scratched and were homogenized in an ice-cold cell lysis buffer (50.0 mmol/L Tris pH=7.6, 150.0 mmol/L NaCl, 0.1% sodium dodecyl sulfate (SDS), 1.0% NP-40). The lysate was centrifuged at 15000×g for 15 min at 4°C and the cellular supernatant was collected. Thirty micrograms protein samples per hole were separated with 15% SDS-polyacrylamide gel electrophoresis (PAGE) gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, MA, U.S.A.). After the blockage of non-specific binding sites with 5% defatted milk solution at room temperature for 1 h, the membrane was incubated overnight 4°C with the anti-caspase 3 antibody. The anti-caspase 3 antibody from Cell Signaling Technology detects endogenous levels of full length of caspase 3 (35 kDa) and the large fragment of caspase 3 resulted from cleavage (17 kDa).
RNA PreparationTotal RNA was extracted using the Trizol Reagent (Invitrogen, Carlsbad, CA, U.S.A.) following H2O2 treatment for 24 h. The samples from each group consisted of an equal amount of total RNA from three replicates to normalize the individual differences. The RNA purity and quality were assessed with a UV spectrophotometer K5500 (Kaiao, Beijing, China), agarose gel electrophoresis and the Agilent 2200 Bioanalyzer (Agilent, U.S.A.). Samples with A260/A280≥1.5, A260/A230≥1, RNA integrity number ≥7 were subjected to DNA microarray.
DNA Microarray and DNA Chip Data AnalysisWe have set up three groups: normal group (normal), hydrogen peroxide group (H2O2), pre-protected with AF-HF001 in hydrogen peroxide group (AF-HF001+H2O2). Each group prepared three separate batches. RNA was extracted and mixed three batches in each group after ensuring RNA quality. One microgram of total RNA was reverse transcribed into cRNA with the Amino Allyl messageAmp™ II kit (Invitrogen), and 8 µg of cRNA was labeled using Amino Allyl MessageAmp™ II cRNA with the Cy5 Kit (Invitrogen). RiboArray™ arrays (Ribo, Guangzhou, China) were hybridized containing 29659 rat genes according to the Ribo Gene Hybridization Protocol. After pre-hybridization, hybridization with the chip was performed at 40°C for 16 h, and the chip was subsequently washed with buffers (Ribo). The microarray was loaded, and the GenePix 4000B scanner and GenePix Pro7 software (Molecular Devices Inc., Sunnyvale, CA, U.S.A.) were used to scan and analyze the array. The quantile normalization method was used to preprocess the microarray data. All of the microarray analyses, including the preprocessing, normalization and statistical analysis, were performed in R-3.0.2. The fold change is the ratio of the normalized spot intensities between the test sample and the control sample. The standard selection criteria to identify the differentially expressed genes are established at |Fold Change|≥1.5.
Statistical AnalysisAll of the experimental data represent the mean of at least three separate experiments and are expressed as the mean±standard deviation (S.D.). A p-value <0.05 was considered statistically significant.
Oxidative stress during hypoxia and ischemia–reperfusion (I/R) injury induces H9c2 apoptosis. As shown in Fig. 2, when H9c2 cells in the experimental groups were subjected to 100 µM H2O2 (Figs. 2A, C) or 200 µM CoCl2 treatment (Figs. 2B, D), the level of caspase 3 activation and DNA fragmentation was greatly increased compared with that of the controls. However, the cytotoxicity of H2O2 and CoCl2 was attenuated with the incubation of AF-HF001. In addition, pretreatment with various concentrations of AF-HF001 (25, 50, 100, or 250 nM) decreased the amount of cleaved caspase 3 in a dose-dependent manner. In addition, AF-HF001 delayed the production of cleaved caspase 3 in a dose-dependent manner in normal cells without oxidative stimuli (Fig. 2E).
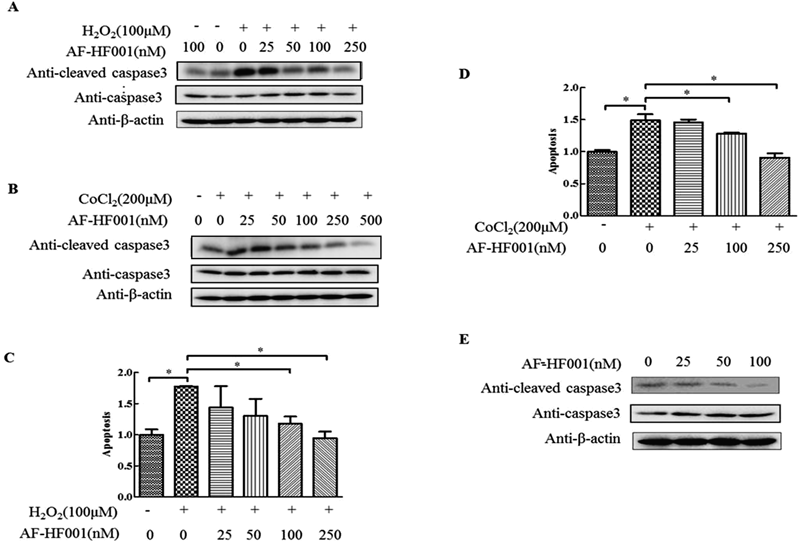
Caspase 3 activation in H9c2 cells exposed to 25 to 250 nM (25 to 500 nM) AF-HF001 for 12 h followed by 100 µM H2O2 (A) or 200 µM CoCl2 (B) for 24 h. H9c2 cell apoptosis in the different groups was evaluated by calculating the absorbance values in a DNA ladder ELISA (C) and (D). (E) Caspase 3 activation in H9c2 cells exposed to 25 to 100 nM AF-HF001 without H2O2. Significance is indicated as * p<0.05.
Previous reports have demonstrated that H2O2 and CoCl2 promote the accumulation of ROS, which act as a second messenger to activate intracellular signaling pathways involved in cardiomyocyte apoptosis.4,5) We then examined ROS production using DCF-DA, which is sensitive to ROS. As illustrated in Fig. 3, H2O2 treatment alone caused strong DCF-DA staining, whereas H2O2 treatment with AF-HF001 produced relatively dim staining in the H9c2 cells. Preincubation with AF-HF001 prior to H2O2 treatment reduced the staining density more effectively than the control.

ROS levels were assessed by DCF-DA assays. Scale bars indicate 50 µm.
We have set up three groups: normal group (normal), hydrogen peroxide group (H2O2), pre-protected with AF-HF001 in hydrogen peroxide group (AF-HF001+H2O2). Each group prepared three separate batches. RNA was extracted and mixed three batches in each group after ensuring RNA quality. All of the RNA samples subjected to DNA microarray exhibited A260/A280≥1.8 and A260/A230≥2.0. In addition, the RNA integrity number was ≥8.1. DNA microarray was performed, and the gene transcript abundance in the cells was compared as follows: incubation with H2O2 medium (H2O2) versus regular medium (control) revealed the effect of exposure to H2O2 alone; incubation with H2O2 medium with AF-HF001 preincubation (AF-HF001+H2O2) versus H2O2 medium (H2O2) revealed the effect generated by AF-HF001 under conditions of oxidative stress alone. Differentially expressed genes were identified with |Fold Change|≥1.5.
The gene expression results for cells treated with H2O2 versus control as well as AF-HF001+H2O2 versus H2O2 were clustered separately in Fig. 4A, which demonstrated that the alteration of gene expression patterns differed in these two groups of comparison. As shown in Fig. 4B, H2O2 resulted in 1658 genes up-regulated by at least 1.5-fold and 2685 genes down-regulated by at least 1.5-fold, whereas the numbers of genes up- /down-regulated by at least 1.5-fold in AF-HF001+H2O2 compared with H2O2 were 2282 and 1882, respectively. In total, 911 shared genes were up-regulated in AF-HF001+H2O2, whereas these genes were down-regulated in H2O2 alone. In addition, 411 shared genes were up-regulated in H2O2 alone, whereas these genes were down-regulated in AF-HF001+H2O2.
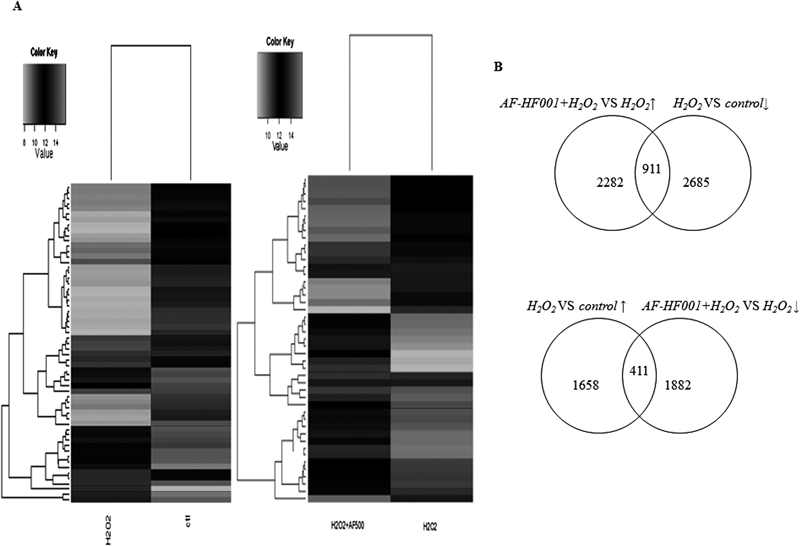
(A) The hierarchical cluster was created from genes based on the threshold of |Fold change|≥4 and two comparisons: H2O2 versus control and AF-HF001+H2O2 versus H2O2. The color key indicates the expression level of the genes exhibiting fold changes: black represents relatively high expression, whereas white represents relatively low expression. (B) Numbers of genes up-regulated or down-regulated by at least 1.5-fold in each group (H2O2 versus control and AF-HF001+H2O2 versus H2O2) and numbers of genes shared in each two groups.
We found 39 genes in H2O2 and 23 genes in AF-HF001+H2O2 that modified their expression with a ratio ≥4.0 for up-regulated genes and a ratio ≤−4.0 for down-regulated genes (Tables 1, 2). Thus, gene ontology analysis was applied to further analyze the microarray data. Among these genes, the shared genes are related to several molecular functions, including phosphatase activity, kinase activity, transferase activity, receptor activity, enzyme activator activity, protein and transcription factor binding, ribosome structural constituent, and biological processes, including metabolism, transport, translation, biosynthesis, complex assembly, cellular localization, signal transduction, cell–cell communication, and apoptosis.
| AF-HF001+H2O2 versus H2O2 (911/2282+411/1882 genes) | |||
|---|---|---|---|
| ID | Name | Ratio | Biological process |
| RB_p_0000218086 | XM_002729221:Rps12l3* | 21.41 | Translation, metabolic process, gene expression, biosynthetic process |
| RB_p_0000229306 | NM_017294:Pacsin1* | 15.21 | Cellular component organization or biogenesis, negative regulation of endocytosis |
| RB_p_0000207974 | NM_053366:Rab6a* | 13.68 | Metabolic process, cellular localization, regulation of catalytic activity, intracellular transport, Ras protein signal transduction |
| RB_p_0000209961 | XM_001055938:Zfp353* | 9.48 | |
| RB_p_0000209572 | XM_002725338:LOC685669* | 5.32 | |
| RB_p_0000208604 | NM_001127298:Wdr91@ | 5.27 | |
| RB_p_0000207861 | NM_001136183:Hsbp1l1@ | 4.67 | |
| RB_p_0000220065 | XM_003750845:LOC100912362* | 4.48 | |
| RB_p_0000209877 | XM_001063134:LOC682793@ | 4.34 | |
| RB_p_0000218190 | XM_002729250:LOC685619* | 4.23 | |
| RB_p_0000216983 | XM_003748856:Ceacam18* | −9.09 | |
| RB_p_0000210962 | XM_576285:Rps19l1@ | −8.65 | Translation, metabolic process, biosynthetic process |
| RB_p_0000218949 | XM_003750132:LOC690073* | −7.61 | |
| RB_p_0000208290 | NM_001162408:Dusp13* | −5.83 | Metabolic process, protein dephosphorylation |
| RB_p_0000223392 | NM_001271294:Thoc7@ | −5.55 | Cellular localization, intracellular transport, nuclear export |
| RB_p_0000210689 | XM_227932:Olr1729* | −5.25 | Multicellular organismal process |
| RB_p_0000228790 | NM_013190:Pfkl@ | −4.80 | Cellular metabolic process, cellular localization, cellular component organization or biogenesis, secretion, cell–cell signaling, catabolic process, glycolysis |
| RB_p_0000225480 | NM_053682:Yme1l1@ | −4.72 | Metabolic process, catabolic process, proteolysis |
| RB_p_0000216798 | XM_003748736:LOC682526@ | −4.25 | |
| RB_p_0000202029 | NM_001100977:Mtfr1* | −4.18 | |
| RB_p_0000205128 | NM_001109258:Tmem70@ | −4.16 | Cellular component organization or biogenesis, organelle organization, proton–transporting ATP synthase complex assembly and biogenesis |
| RB_p_0000229286 | NM_017250:Htr2b@ | −4.13 | Metabolic process, biosynthetic process, cellular homeostasis, apoptosis |
| RB_p_0000211999 | NM_176080:Naglt1@ | −4.11 | Establishment of localization, transmembrane transport |
* Shared genes with 4-fold change in expression. @ Shared genes with 1.5-fold change in expression.
| H2O2 versus control (411/1658+911/2685 genes) | |||
|---|---|---|---|
| ID | Name | Ratio | Molecular function |
| RB_p_0000211180 | NM_001177904:LOC100363776@ | 8.71 | |
| RB_p_0000216983 | XM_003748856:Ceacam18* | 7.38 | |
| RB_p_0000202029 | NM_001100977:Mtfr1* | 6.88 | |
| RB_p_0000218949 | XM_003750132:LOC690073* | 6.60 | |
| RB_p_0000208290 | NM_001162408:Dusp13* | 4.66 | Protein tyrosine phosphatase activity |
| RB_p_0000210689 | XM_227932:Olr1729* | 4.39 | G-Protein coupled receptor activity |
| RB_p_0000222110 | XM_003752113:Arxes2@ | 4.23 | |
| RB_p_0000220432 | XM_002727894:LOC100359498@ | −46.90 | |
| RB_p_0000210854 | XM_002729407:LOC688513@ | −39.70 | |
| RB_p_0000224599 | NM_001000614:Olr756@ | −35.28 | G-Protein coupled receptor activity |
| RB_p_0000218086 | XM_002729221:Rps12l3* | −21.65 | Structural constituent of ribosome |
| RB_p_0000218764 | XM_003750014:LOC679803@ | −18.12 | |
| RB_p_0000205490 | NM_012882:Sstr5@ | −17.17 | Binding, peptide receptor activity |
| RB_p_0000209961 | XM_001055938:Zfp353* | −16.82 | |
| RB_p_0000207974 | NM_053366:Rab6a* | −14.57 | Binding, enzyme activator activity |
| RB_p_0000219019 | XM_003750184:LOC690335@ | −12.24 | |
| RB_p_0000214561 | XM_001056364:RGD1566102@ | −11.52 | |
| RB_p_0000214135 | XM_003753315:LOC100912860@ | −11.49 | |
| RB_p_0000221098 | XM_001080534:Esco2@ | −10.53 | |
| RB_p_0000200783 | NM_022384:Ascl1@ | −10.51 | Protein binding, transcription factor binding transcription factor activity |
| RB_p_0000229306 | NM_017294:Pacsin1* | −10.36 | Cytoskeletal protein binding, protein binding |
| RB_p_0000206375 | NM_080787:Dgka@ | −8.45 | Binding, transferase activity, diacylglycerol kinase activity |
| RB_p_0000202037 | NM_001100987:Aqr@ | −8.28 | |
| RB_p_0000203710 | NM_001106196:Galnt2@ | −7.98 | Binding, transferase activity |
| RB_p_0000210333 | XM_237757:Ccdc154@ | −7.18 | |
| RB_p_0000209559 | XM_001069890:LOC684327@ | −7.07 | |
| RB_p_0000205478 | NM_012649:Sdc4@ | −6.45 | Protein binding, thrombospondin receptor activity |
| RB_p_0000209572 | XM_002725338:LOC685669* | −6.00 | |
| RB_p_0000204592 | NM_001109458:Fam163b@ | −5.68 | |
| RB_p_0000218190 | XM_002729250:LOC685619* | −5.64 | Protein tyrosine kinase activity, binding, purine ribonucleotide |
| RB_p_0000216055 | XM_003754474:LOC367140@ | −5.57 | |
| RB_p_0000225393 | NM_001004209:Hsd17b11@ | −5.05 | Binding, steroid dehydrogenase activity, catalytic activity, oxidoreductase activity |
| RB_p_0000219746 | XM_003750644:LOC100911599@ | −4.78 | |
| RB_p_0000213139 | XM_001069205:LOC684158@ | −4.59 | |
| RB_p_0000200844 | NM_001079706:Exnef@ | −4.39 | Hydrolase activity, acting on ester bonds |
| RB_p_0000220065 | XM_003750845:LOC100912362* | −4.34 | |
| RB_p_0000206820 | NM_133390:Zfp347@ | −4.22 | |
| RB_p_0000203791 | NM_001108884:Psma8@ | −4.04 | Hydrolase activity, peptidase activity |
| RB_p_0000218926 | XM_003750115:LOC100362369@ | −4.01 | |
* Shared genes with 4-fold change in expression. @ Shared genes with 1.5-fold change in expression.
Due to its high morbidity and mortality, heart failure has been a public health burden worldwide. AF-HF001 could be a potential therapeutic candidate for the treatment of heart failure. Here, we analyzed differences in the apoptosis level and gene expression in H2O2-treated cardiomyocytes cultured with or without pretreated AF-HF001. We aimed to identify genes that contribute to the protective effects of AF-HF001 and provide new insights in the potential use of AF-HF001 as a treatment for heart failure treatment.
The protection of cardiomyocytes from ROS-induced apoptosis may be a beneficial therapeutic intervention to combat heart failure. The data reported in this study demonstrated that AF-HF001 protects H9c2 cells from hypoxia-induced injury and attenuates ROS production. Moreover, AF-HF001 exhibited potential in prolonging normal cell viability, which is a significant benefit for cardiomyocytes, as the mature human heart was reported to have a detectable but low regenerative capacity.6,7) It is conceivable that AF-HF001 may exhibit therapeutic utility in the prevention of heart failure.
As a metabolically active organ, high ROS levels exist in the cardiac tissue. Once an imbalance arises in the ROS generation and the antioxidant system, an overload of ROS induces oxidative stress, leading to cell dysfunction. Mitochondrial complex I (reduced nicotinamide adenine dinucleotide (NADH) ubiquinone oxidoreductase) initiates the mitochondrial electron transport chain, and a decrease in the activity of mitochondrial complex I is associated with an increase in ROS production.8) Jacquemin et al.9) modified the mitochondria by cleaving the Ndufs2 subunit to trigger ROS-dependent apoptosis. Massoz et al.10) reported that Ndufs3 was required for complex I activity, and Ndufs3 RNA interference reduced intracellular ROS production. In this study, AF-HF001 reversed the down-regulation of Ndufs2 (1.53-fold), Ndufb6 (1.62-fold), Ndufb8 (1.59-fold), and Ndufa13 (1.69-fold) as well as the up-regulation of Ndufs3 (−2.10-fold) and Ndufs5 (−2.18-fold). These genes are structural subunits of mitochondrial complex I. Although the significance of the alteration of these subunits in heart failure remains unclear, these changes may be associated with ROS production and pathological progression. Thus, we suggest that AF-HF001 acts as an ROS scavenger by altering the expression of the Nduf (NADH dehydrogenase (ubiquinone) 1) genes.
Impaired contractility is the primary phenotype of heart failure. Cardiac sarcomere proteins consist of contractile proteins that govern the mechanical contractility process and cytoskeleton proteins that regulate the cardiomyocyte scaffold. Hershberger et al. reported that patients with familial or idiopathic dilated cardiomyopathy express rare variants of five genes, including myosin binding protein C (MYBPC3), Į-myosin heavy chain (MYH6), tropomyosin 1 (TPM1), troponin C (TNNC1) and cardiac troponin I (TNNI3).11) Tpm1 is classified as tropomyosin and stabilizes actin filaments and cables, and the unacetylated form of Tpm1 significantly reduces the actin-binding activity.12) The MYH14 protein was detected in rat and mouse heart and represents a slow-type myosin that acts as the molecular motor to drive contraction.13) An autosomal dominant mutation in MYH14 (myosin, heavy chain 14, non-muscle) was reported to be associated with myopathy.14) In our experiments, we found that AF-HF001 treatment decreased the expression of Tpm1 (−2.76-fold, tropomyosin 1, alpha) and MYH14 (−1.61-fold) in H2O2-treated H9c2 cells.
The proteins associated with contractility are involved in calcium ion (Ca2+)-dependent excitation–contraction coupling. The ryanodine receptor (RyR) is a key protein in the regulation of intracellular Ca2+ release, and the amount and activity of RyR decreases in failing animal myocardium.15) In our microarray data, H9c2 cells responded to H2O2 by increasing RyR1 (1.6-fold), whereas AF-HF001 restored the normal levels of expression (−1.57-fold). Altogether, AF-HF001 may avoid the negative effect on sarcomere proteins and retain the balance of Ca2+ storage and release to preserve cardiac contractility.
The extracellular matrix is a complex microenvironment that includes a large number of matrix proteins, signaling proteins, proteases, etc. Structural remodeling of the extracellular matrix is an important contributor to abnormal ventricular function and depends on the balance of matrix metalloprotease and tissue inhibitors of matrix metalloprotease (TIMP).16) TIMP4 inhibits MMP-mediated degradation of excess extracellular matrix and reduces myocardial infarction in vivo in I/R mice.17) Chaturvedi et al.18) reported that TIMP4 attenuates oxidative stress and induces the differentiation of cardiac progenitor cells into cardiomyocytes. Our data demonstrated that TIMP4 expression upon H2O2 treatment (−1.85-fold) was up-regulated in cells that were also treated with AF-HF001 (1.97-fold). Thus, AF-HF001 may participate in extracellular matrix remodeling.
As is stated above, we investigated the relationship between gene expression and heart disease and suggested a molecular mechanism by which AF-HF001 protects heart tissue that is related to ROS generation, cardiac contractility and extracellular matrix remodeling. Furthermore, AF-HF001 affects several signaling pathways that are involved in the response to oxidative stress.
In response to ROS, MAP kinase family members play an important role in mediating signaling pathways. AF-HF001 induced increased expression of the A-Raf proto-oncogene, serine/threonine kinase (A-Raf, 1.80-fold), which belongs to the Raf family of protein serine/threonine kinases. A-Raf acts as a scaffold protein to stabilize B-Raf : C-Raf complexes.19) In addition, three Raf family members are all expressed in heart tissue. Raf family members possess serine/threonine kinase activity and activate the mitogen-activated protein kinase kinases (MKKs)/extracellular signal-regulated kinases (ERKs) pathway to regulate cardiomyocyte survival and growth.20) Additionally, AF-HF001 increased the expression of mitogen-activated protein kinase kinase 5 (MAP2K5, 1.54-fold) compared with the H2O2 medium (−1.81-fold). MAP2K5 is down-regulated in apoptosis as previously reported, and its expression regulates cell survival.21) Several studies have revealed that MAP2K5 phosphorylates ERK5 downstream and that the MAP2K5/ERK5 pathway regulates cardiac hypertrophy.22) Our data indicate that Dusp13A (dual specificity phosphatase 13a) is down-regulated (−5.83-fold) by AF-HF001 but up-regulated (4.66-fold) by H2O2. Dusp13A physically interacts with and activates ASK1, which is a MAP kinase kinase kinase, leading to the initiation of the c-Jun N-terminal kinase (JNK)/p38-mediated apoptosis pathway.23) The study on functional regulation between MAPK and MAPK kinase is in progress and the result should be reported in the near future.
The intrinsic mitochondrial pathway is one of the apoptotic pathways that exist in cells. The genes that encode the components of the intrinsic mitochondrial pathway, such as Bax, Hsp27 (Hspb1), Apaf1, casp3 and casp7, were significantly down-regulated −1.52-fold, −1.65-fold, −3.42-fold, −1.74-fold and −1.63 fold, respectively, by AF-HF001 compared with H2O2. Bcl2l1 was up-regulated 1.93-fold upon AF-HF001 treatment. Bax translocates from the cytosol to the mitochondria in response to apoptotic stimuli, and it oligomerizes to form pores in the outer mitochondrial membrane; these pores allow cytotoxic proteins to cross from the intermembrane space into the cytosol to induce apoptosis.24) Apoptotic protease-activating factor 1 (APAF1) binds to cytochrome c and forms the apoptosome, which recruits and activates caspase 9.25) Subsequently, caspases 3 and 7 are cleaved by caspase 9. Caspase 3 activation may be dependent on or independent of caspase 9 and acts as a critical cell killer or death inducer.26) Bcl2l1 is a member of the antiapoptotic Bcl-2 family, preserving mitochondrial integrity under normal conditions by inhibiting the activation of the proapoptotic Bcl-2 proteins; ERK/c-Jun mediates its expression.27) Considering the results described above, we propose that AF-HF001 inhibits apoptosis by regulating the intrinsic mitochondrial pathway.
In conclusion, AF-HF001 reverses changes in gene expression induced by oxidative damage in H9c2 cardiomyocytes and has a protective effect on these cells. It possesses potential as a candidate for the treatment of heart failure.
This work was supported by the National Natural Science Foundation of China (81370683) and Prospective joint research project in Jiangsu Province (BY2015019-25).
The authors declare no conflict of interest.