2016 Volume 39 Issue 4 Pages 492-501
2016 Volume 39 Issue 4 Pages 492-501
Terminalia, a large genus of Combretaceae, is distributed in Tropical Asia, Africa, and America. Some Terminalia plants are used in folk medicine because they possess powerful medicinal properties. Dried fruits of Terminalia bellirica and Terminalia chebula are used as the main ingredient in Triphala, a famous polyherbal formulation in Ayurvedic medicine and Thai folk medicine, because of their laxative, detoxifying, and rejuvenating effects. To clarify the phylogenetic relationships of medicinal Terminalia species (T. bellirica, T. chebula, and T. catappa) and authenticate their crude drugs, “Samo” and Triphala, nucleotide sequencing alignments in the internal transcribed spacer one–two (ITS 1–2) regions of Terminalia plants collected in Thailand were performed. The amplified fragments of Terminalia species were approximately 800 bp in length. To compare these sequences and DDBJ registered data, a molecular phylogenetic tree was constructed. Phylogenetic analysis clearly separated the sequences into two groups: Asian Terminalia and African Terminalia with some exceptions. In the analyzed sequences, the length of the ITS1-5.8S-ITS2 region was 674 bp in T. chebula, and 677 bp in T. bellirica and T. catappa. Eighty-one single nucleotide polymorphisms (SNPs) and nine insertion–deletions (indels) were observed, and the nucleotide sequences of this region showed species-specific sequences. Based on these differences, polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) and amplification refractory mutation system (ARMS) were applied to identify medicinal Terminalia species. Moreover, the ARMS method was chosen for fingerprinting analysis of Samo crude drugs and Triphala formulations because it was a fast, cost-effective, and reproducible approach.
Terminalia, a large genus of Combretaceae, is distributed in Tropical Asia, Africa, and America.1) It is widely used as a medicinal plant in folk medicine. Of the Terminalia species, T. arjuna (DC.) WIGHT & ARN., T. bellirica (GAERTN.) ROXB., T. chebula RETZ., and T. catappa L. exhibit potent medicinal properties. In Thailand, five of seventeen Terminalia plants, T. chebula RETZ. var. chebula, T. chebula RETZ. var. nana GAGNEP., T. bellirica (GAERTN.) ROXB., T. citrina (GAERTN.) ROXB. ex FLEMING, and T. catappa L., are used in Thai traditional medicine.2) Moreover, two of the three main ingredients in Triphala are composed of T. chebula var. chebula, T. bellirica, and Phyllanthus emblica (Euphorbiaceae) fruit powder. Triphala, one of the most important and well-known multi-component formulations in Ayurvedic medicine and Thai folk medicine, exhibit laxative, detoxifying, and rejuvenating effects.3) Pharmacological studies have shown that Triphala extract possesses anticancer,4) immunomodulatory,5) radioprotective,6) antioxidant,7,8) anti-inflammatory,9) and hypolipidemic activities.10)
Anatomical and chemical analyses, conventional tools for the authentication of herbal medicine, are frequently used to identify the botanical origin of herbal medicine. However, both methods have limitations. For example, it is very difficult to identify the botanical origin on the basis of processed herbal materials (mixed powder form or shredded pieces).11,12) Furthermore, chemical identification may be affected by physiological condition, growth condition, harvesting period, post-harvest processing, sample freshness, and storage condition.11,13,14) Species identification at the DNA level is an alternative tool for the authentication of morphologically ambiguous herbal drugs.11) In recent years, DNA analysis has been utilized as a powerful method for the identification of medicinal plants.15)
Single nucleotide polymorphisms (SNPs) are DNA variations in plants and animals, and are used as high-quality molecular markers for practically distinguishing single base differences within the genome.16,17) Hybridization, primer extension, oligonucleotide ligation, allele-specific polymerase chain reaction (PCR), and endonuclease cleavage are utilized for SNP genotyping.18) PCR-restriction fragment length polymorphism (RFLP) is a simple and reliable method that uses a restriction endonuclease that has high affinity for unique restriction sites for species authentication.19–23) The amplification refractory mutation system (ARMS), an easy and cost-effective method for authenticating herbs and their adulterants, is an allele-specific PCR that allows discrimination of alleles at specific loci differing by as little as 1 bp.24) Both techniques are based on the generic variation of SNPs.
It is very difficult to authenticate Triphala (a mixture of fruit powders) by morphological and chemical methods. Because of that, a molecular technique is introduced to identify the ingredients. Previously, Dnyaneshwar et al. developed the randomly amplified polymorphic DNA (RAPD) based sequence characterized amplified region (SCAR) marker technique and applied it to the identification of P. emblica.25) Therefore, all ingredients in Triphala should be identified. In the present study, the PCR-RFLP and ARMS methods were used to compare the efficacy in the internal transcribed spacer (ITS) region of nuclear ribosomal DNA (nrDNA) in order to select an accurate and convenient method for the authentication of all ingredients in Triphala. In addition, the molecular phylogenetic relationships of Terminalia species were analyzed on the basis of the nucleotide sequence alignments of these plant specimens, including the sequences retrieved from DNA database.
Leaf samples of nine Terminalia species, five of which are medicinal Terminalia species, namely, T. bellirica (GAERTN.) ROXB., T. chebula RETZ. var. chebula, T. chebula RETZ. var. nana GAGNEP., T. catappa L., and T. citrina (GAERTN.) ROXB. ex FLEMING were collected from Thailand. In addition, P. emblica L. and Combretum indicum (L.) DEFILIPPS were collected for use as nucleotide sequence reference and in phylogenetic tree construction, respectively. The leaf specimens were identified by Dr. Monthon Norsaengsri, botanist of Queen Sirikit Botanic Garden, Thailand. Voucher samples were deposited in Queen Sirikit Botanic Garden Herbarium (QBG), Chiang Mai, Thailand and the Herbarium of the Laboratory of Molecular Pharmacognosy of the Graduate School of Medical Science, Kanazawa University, Japan (Table 1). Three crude drug samples (Samo Thai, Samo Phiphek, and Makampom) and nine commercial Triphala formulations were obtained from a local market in Thailand (Table 2).
| Species | No. of samples | Voucher no. | Collection site | DDBJ/EMBL/GenBank accession no. |
|---|---|---|---|---|
| T. bellirica (GAERTN.) ROXB. | 10 | B13090711-2 | Sanam Chai Khet, Chacheongsao | LC050567 |
| B130909221 | Maerim, Chiang Mai | |||
| B130910241 | Muang, Chiang Mai | |||
| B130912611-3 | Ban Tak, Tak | |||
| B130917221 | Muang, Chiang Mai | |||
| B130917271-2 | Sansai, Chiang Mai | |||
| T. chebula RETZ. var. chebula | 17 | C130908311-2 | Muang, Lamphun | LC050565 |
| C130914311-2 | ||||
| CN130908321 | ||||
| CN130914311 | ||||
| C130909221 | Maerim, Chiang Mai | |||
| C130910231-2, C130910241 | Muang, Chiang Mai | |||
| C130911511 | Muang, Udon Thani | |||
| C130912611-3 | Ban Tak, Tak | |||
| C130915271-2 | Sansai, Chiang Mai | |||
| C130917281 | Muang, Lampang | |||
| T. chebula RETZ. var. nana GAGNEP. | 1 | CN130916221 | Maerim, Chiang Mai | LC050566 |
| T. catappa L. | 7 | D130910241-5 | Muang, Chiang Mai | LC050568 |
| D130911511 | Muang, Udon Thani | |||
| D130913261 | Maerim, Chiang Mai | |||
| T. citrina (GAERTN.) ROXB. ex FLEMING | 3 | R130918811 | Phra Nakhon, Bangkok | LC050564 |
| R140630811 | ||||
| R140630821 | ||||
| T. mantaly H. PERRIER | 7 | I130908211 | Sankampaeng, Chiang Mai | LC050569 |
| I130908331-2 | Muang, Chiang Mai | |||
| I130910241 | Muang, Chiang Mai | |||
| I130912621 | Ban Tak, Tak | |||
| I130913261-2 | Maerim, Chiang Mai | |||
| T. glaucifolia CRAIB | 1 | K130909221 | Maerim, Chiang Mai | LC050562 |
| T. elliptica WILLD. | 5 | A130909411-2 | Mae Sariang, Mae Hong Son | LC050570 |
| A130911511 | Muang, Udon Thani | |||
| A130912611-2 | Ban Tak, Tak | |||
| T. mucronata CRAIB & HUTCH | 7 | M130912611-3 | Ban Tak, Tak | LC050563 |
| G130912611-3 | Ban Tak, Tak | |||
| M130917711 | Muang, Lampang | |||
| Combretum indicum (L.) DEFILIPPS (Combretaceae) | 2 | L130917711 | Muang, Lampang | LC050571 |
| L130918241 | Muang, Chiang Mai | |||
| Phyllanthus. emblica L. (Euphorbiaceae) | 11 | P140605211-2 | Maerim, Chiang Mai | LC089029 |
| P140605221-2 | Muang, Chiang Mai | |||
| P140605661-2 | Ban Tak, Tak | |||
| P140605231-2 | Mae Chaem, Chiang Mai | |||
| P140605241-2 | Omkoi, Chiang Mai | |||
| P140605251 | Hot, Chiang Mai |
| Crude drug name | Drug origina) | Sample ID | Collection date | Collection site |
|---|---|---|---|---|
| Samo Thai | T. chebula | DF 2, 6 | 11 Sep 2013 | Muang, Udon Thani |
| DF 12 | 13 Sep 2013 | Muang, Tak | ||
| DF 17, 22, 24 | 7 Sep 2013 | Samphanthawong, Bangkok | ||
| DF 28 | 14 Sep 2013 | Muang, Chiang Mai | ||
| DF 32–33 | 17 Sep 2013 | Muang, Lampang | ||
| DF 38 | 10 Oct 2013 | Hat Yai, Songkhla | ||
| Samo Phiphek | T. bellirica | DF 3, 5, 10 | 11 Sep 2013 | Muang, Udon Thani |
| DF 13 | 13 Sep 2013 | Muang, Tak | ||
| DF 18, 23, 25 | 7 Sep 2013 | Samphanthawong, Bangkok | ||
| DF 30 | 14 Sep 2013 | Muang, Chiang Mai | ||
| DF 34–35 | 17 Sep 2013 | Muang, Lampang | ||
| DF 40 | 10 Oct 2013 | Hat Yai, Songkhla | ||
| Makampom | P. emblica | DF 4, 7–8 | 11 Sep 2013 | Muang, Udon Thani |
| DF 15 | 13 Sep 2013 | Muang, Tak | ||
| DF 20, 26 | 7 Sep 2013 | Samphanthawong, Bangkok | ||
| DF 31 | 14 Sep 2013 | Muang, Chiang Mai | ||
| DF 36–37 | 17 Sep 2013 | Muang, Lampang | ||
| DF 42 | 10 Oct 2013 | Hat Yai, Songkhla | ||
| Triphala formulation | T. chebula : T. bellirica : P. emblica (1 : 1 : 1) | TP 1 | 17 Sep 2013 | Muang, Lampang |
| TP 2 | 7 Sep 2013 | Bang Krathum, Phitsanulok | ||
| TP 3 | 7 Sep 2013 | Sampran, Nakornpathom | ||
| TP 4 | 7 Sep 2013 | Muang, Prachin Buri | ||
| TP 5–7 | 7 Sep 2013 | Samphanthawong, Bangkok | ||
| TP 8 | 11 Sep 2013 | Muang, Udon Thani | ||
| TP 9 | 14 Sep 2013 | Muang, Chiang Mai |
a) Origin was identified by sequence alignments of ITS region.
Total DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol with minor modifications.
PCR AmplificationNuclear internal transcribed spacer (ITS) was amplified by PCR using 100–120 ng of total DNA as the template in 25 µL of a reaction mixture containing 12.5 µL of 2× PCR Buffer for KOD FX Neo, 0.4 mM of each deoxyribonucleotide triphosphate (dNTP), 0.15 µM of each primer (Ter.Af+Un.3R for ITS1, and Un.3F+Ake-26SR for ITS2 in Table 3), and 0.5 unit of KOD FX Neo DNA polymerase (Toyobo, Japan). PCR amplification was performed under the following cycling conditions: hot start at 94°C for 2 min; 30 cycles of denaturation at 94°C for 15 s, annealing at 62°C for 30 s, and elongation at 68°C for 45 s; and final elongation at 68°C for 5 min. The PCR product was purified with a Fast Gene™ Gel/PCR Extraction Kit (Nippon Genetics Europe GmbH, Japan).
| Primer name | Sequence (5′→3′) | Length (bp) | Tm (°C) |
|---|---|---|---|
| Ter.Af | CGA GAA GTC CAC TGA ACC TT | 20 | 60 |
| Ake-26SR | GTA AGT TTC TTC TCC TCC GC | 20 | 60 |
| Un.3F | CGA CTC TCG GCA AGG GAT AT | 20 | 65 |
| Un.3R | AAC TTG CGT TCA AAG ACT CG | 20 | 60 |
| PHE.Bf | CCT TGT GCA CCT GAA GCC A | 19 | 58 |
| PHE.Br | TTC GGC CAA ATG AAC GAG G | 19 | 60 |
| TCA.Af | CGT TTT TTA AAT GCC CGG GAT A | 22 | 62 |
| TCH.Af | AGC GCC AAG GTA CTC CAA CAA | 22 | 68 |
| TBE.Cf | GGG CTG CTG TTC AAC GTC ATA AT | 23 | 68 |
| Ter.Br | GAT CTG GAG GCA ACG CGA | 18 | 58 |
The purified PCR fragment was subjected to direct sequencing with a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) using an ABI PRISM 310 sequencer (Applied Biosystems). The obtained DNA information was aligned using ‘DNASIS’ version 3 (Hitachi, Japan).
Phylogenetic AnalysisThe aligned DNA sequences of the ITS1-5.8S-ITS2 region were analyzed using Molecular Evolutionary Genetic Analysis (MEGA) version 5.2.2 Software.26) Maximum likelihood calculation was carried out using the Kimura two-parameter model with 1000 bootstrap replications. Apart from the nucleotide sequences of the nine Terminalia species distributed in Thailand as determined in our present study, 26 Terminalia samples from DDBJ/EMBL/GenBank were used for phylogenetic tree reconstruction. C. indicum (L.) DEFILIPPS of the same tribe Combreteae in Family Combretaceae was used as outgroup species for phylogenetic tree rooting.27)
PCR-RFLP AnalysisTer.Af (forward primer) and Ake-26SR (reverse primer) were used for PCR amplification of approximately 800 bp in the ITS1-5.8S-ITS2 region. The purified PCR products from T. catappa, T. chebula var. chebula, and T. bellirica were digested with 10 units of restriction enzyme XspI (TaKaRa Bio, Inc., Japan) at 37°C for 2 h to distinguish T. catappa from the other species. For the identification of T. chebula var. chebula and T. bellirica, 10 units of restriction enzyme Aor13HI (TaKaRa Bio, Inc.) was added, and digestion was carried out at 55°C for 2 h. For the authentication of the Triphala formulation, PCR-RFLP analysis of P. emblica L. (one of the three ingredients of Triphala) was conducted with both restriction enzymes (Fig. 1A). The digested fragments were separated by 3.0% agarose gel electrophoresis and visualized by staining with GelRed™ Nucleic Acid Gel Stain (Wako Pure Chemical Industries, Ltd., Japan).

A. Restriction sites of XspI and Aor13HI in the rDNA-ITS region. B. Amplification fragment sites of species-specific primers for P. emblica, T. catappa, T. chebula var. chebula, and T. bellirica.
From the nucleotide substitutions at different positions in the ITS region (Fig. 1B), six species-specific primers were designed as follows: TCA.Af and Ter.Br for the identification of T. catappa; TCH.Af and Ter.Br for the identification of T. chebula var. chebula; TBE.Cf and Ter.Br for the identification of T. bellirica, and PHE.Bf and PHE.Br for the identification of P. emblica, and were used in the authentication of Triphala (Table 3). Multiplex-ARMS-PCR amplification was performed by using the DNA templates of all the four species, including crude drugs and Triphala samples with multiple species-specific primers. Twenty-five microliters of the reaction mixture consisted of 12.5 µL of GoTaq® Green Master Mix (Promega, U.S.A.), 0.2 µM species-specific primer, and 100–120 ng of DNA templates of leaf samples. Crude drug and Triphala samples were mixed with 12.5 µL of 2× PCR Buffer for KOD FX Neo, 0.4 mM of each dNTP, 0.15 µM of each three sets of species-specific primers, and 0.5 unit of KOD FX Neo DNA Polymerase (Toyobo, Japan). Amplification was carried out under the following conditions: pre-heating at 94°C for 2 min, followed by 30 cycles at 94°C for 15 s, 60°C for 30 s, and 68°C for 45 s, and a final extension at 68°C for 5 min. The amplified PCR products were detected by 2.0% agarose gel electrophoresis and visualized by staining with GelRed™ Nucleic Acid Gel Stain under UV light irradiation.
The assembled nucleotide sequences of all samples have been deposited in the DDBJ Nucleotide Sequence Database. Nucleotide differences in the ITS1-5.8S-ITS2 regions, where selected nucleotides were compared with each other and the involved nucleotides in medicinal Terminalia species, are summarized in Table 4. All Terminalia species did not show intraspecific variation in each species. The lengths of the ITS1-5.8S-ITS2 regions were 674 bp in T. chebula var. chebula and T. citrina, and 677 bp in T. bellirica and T. catappa, whereas it was 675 bp in T. chebula var. nana. There were 90 variable sites among the five species: 81 sites were SNPs and 9 sites were indels. In the starting 5.8S coding region of T. chebula var. chebula and T. citrina in same individuals, one A base shift deletion was noted as the overlapping peaks at nucleotide no. 279, whereas that in T. chebula var. nana occurred at nucleotide no. 278 in the electropherogram. In the ITS2 region of T. citrina, moreover, one G base shift deletion was found as the overlapping peaks at nucleotide no. 604. Because of the only one base deletion, we were able to separate each alignment from the overlapping peaks in the electropherogram. DNA sequence analysis of T. chebula var. chebula, T. chebula var. nana, and T. citrina revealed that they had some differences at nucleotide nos. 277–486. For example, T. chebula var. nana had two A bases, one Y (T or C) overlapping nucleotide signal, as well as G and A at nucleotide nos. 277–278, 466, and 485–486, respectively, whereas T. chebula var. chebula had two deletions, C, and two R (A or G) overlapping nucleotide signals, respectively. T. citrina had two overlapping nucleotide signals (R and Y) instead of A and T in T. chebula var. chebula at nucleotide nos. 408 and 417, respectively. T. chebula var. chebula had three overlapping nucleotide signals (Y and RR) at nucleotide nos. 451 and 485–486, respectively, whereas T. citrina had T and GA at the same nucleotide numbers.
| Species | Nucleotide numbera) | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS1 | 5.8Sb) | |||||||||||||||||||||||||||||||||||||
| 44–5 | 53 | 60 | 76 | 83–5 | 93–4 | 105 | 108–10 | 112 | 115 | 119 | 125 | 128 | 132 | 140–2 | 150 | 157 | 160–1 | 167 | 169 | 174 | 179 | 194 | 209–11 | 215–7 | 224 | 250 | 267 | 269–72 | 277–9 | 408 | 413 | 417 | ||||||
| T. chebula var. chebula | AT | G | A | T | -CA | CA | T | AGC | - | T | T | - | G | C | CGA | G | T | G- | T | G | T | C | R | TCC | AAC | Y | C | G | TGCG | --A | A | C | T | |||||
| --- | ||||||||||||||||||||||||||||||||||||||
| T. chebula var. nana | ** | * | * | * | -** | ** | * | *** | - | * | * | - | * | * | *** | * | * | *- | * | * | * | * | * | *** | *** | * | * | * | **** | -A* | * | * | * | |||||
| --* | ||||||||||||||||||||||||||||||||||||||
| T. citrina | ** | * | * | * | -** | ** | * | *** | - | * | * | - | * | * | *** | * | * | *- | * | * | * | * | * | *** | *** | * | * | * | **** | --* | R | * | Y | |||||
| --- | ||||||||||||||||||||||||||||||||||||||
| T. bellirica | G* | A | * | C | -TG | TG | * | *** | - | * | Y | G | - | T | T** | A | A | R- | A | * | C | Y | G | MYG | G*T | T | * | A | C*TC | TA* | * | * | * | |||||
| T. catappa | *C | A | G | * | ATG | T* | - | TAT | T | C | * | G | A | * | TAG | A | C | CA | C | A | C | * | G | *** | GGT | T | T | A | CATC | --* | * | T | * | |||||
| Species | Nucleotide number | |||||||||||||||||||||||||||||||||||||
| ITS2 | Length (bp) | |||||||||||||||||||||||||||||||||||||
| 443 | 451 | 460 | 462 | 466 | 472 | 476–7 | 480 | 483 | 485–6 | 489 | 506 | 517 | 519 | 522 | 525 | 527 | 529 | 539 | 543 | 547 | 551 | 555 | 560 | 583 | 588–9 | 600 | 604 | 608 | 610 | 613 | 620 | 622–3 | 628–9 | 634 | 638 | 654 | ||
| T. chebula var. chebula | G | Y | T | C | C | C | AG | A | G | RR | Y | C | T | A | A | A | C | A | C | T | C | G | - | A | C | CG | A | G | C | T | C | A | CM | CC | G | R | R | 674 |
| T. chebula var. nana | * | * | * | * | Y | * | ** | * | * | GA | * | * | * | * | * | * | * | * | * | * | * | * | - | * | * | ** | * | * | * | * | * | * | ** | ** | * | * | * | 675 |
| T. citrinac) | * | T | * | * | * | * | ** | * | * | GA | * | * | * | * | * | * | * | * | * | * | * | * | - | * | * | ** | * | * | * | * | * | * | ** | ** | * | * | * | 674 |
| - | ||||||||||||||||||||||||||||||||||||||
| T. bellirica | A | T | G | T | T | * | *M | T | C | GT | T | * | * | * | G | C | T | G | A | C | T | * | G | G | * | TA | G | * | G | C | T | * | AT | ** | C | A | G | 677 |
| T. catappa | * | T | A | * | T | T | TT | T | C | GT | T | T | C | G | * | * | * | * | * | C | T | A | - | G | T | *A | G | * | G | C | T | G | *T | TT | T | A | A | 677 |
Asterisk (*) indicates the same nucleotide as top sequence; hyphen (-) indicates nucleotide deletion; Y denotes C and T; R denotes A and G; and M denotes A and C. a) The number 1 at the nucleotide position is the first nucleotide of ITS1. b) T. chebula var. chebula and T. citrina have two overlapping alignments from nucleotide number 279 in the same individual, whereas T. chebula var. nana also has two overlapping alignments from nucleotide number 278 in 5.8S coding region. c) T. citrina has two overlapping alignments from nucleotide number 604 (ITS2 region) in the same individual.
From the results, it appeared that the nucleotide sequences in the ITS regions of medicinal Terminalia species were species-specific. T. chebula var. chebula, T. chebula var. nana, and T. citrina contained some different nucleotides each others, but they have many dissimilar nucleotides with T. bellirica and T. catappa. T. chebula var. chebula could be distinguished from T. citrina on the basis of fruit characteristics: T. citrina had an ellipsoid and obscurely 5-angled drupe with 5-angled seeds, whereas T. chebula var. chebula bore a subglobose drupe with rough seeds.2) The fruit of T. citrina was used to treat digestive disorders, similar to T. chebula in Thai traditional medicine and Ayurvedic medicine.2,28) Two varieties of T. chebula were found in Thailand. Krachai et al. reported that the morphological, palynological, and anatomical characteristics of those varieties were similar, and habit and presence or absence of tannin in bundle sheath near the leaf lower epidermis were the key characteristics for distinguishing those two varieties.29)
Phylogenetic AnalysisCombretaceae is distributed in the tropics and some warm temperate zones. It is reported that outcrossing is the primary mode of reproduction in Terminalia.30,31) In many tropical tree species, outcrossing is predominant, leading to high genetic diversity within population.32–34) According to Stace,35) the greatest genetic diversity of Terminalia species is found in Southeast Asia. Because of the morphological complexity inter/intra-species, genus Terminalia is still not clearly classified into subgenera and sections.26) Seventeen Terminalia species were found in Thailand, a country located in Southeast Asia.2) In the present study, we collected nine Terminalia species from Thailand, including two species (T. chebula var. nana and T. mucronata) endemic to Southeast Asia.
The molecular phylogenetic relationship among Terminalia species distributed in Thailand was reconstructed on the basis of the nucleotide sequences in the ITS regions obtained in the present study along with those retrieved from DDBJ/EMBL/GenBank DNA database. Maurin et al.36) analyzed the ITS sequences of Terminalia species collected mainly in Africa as well in Asia, Australia, and the Pacific islands. As shown in Fig. 2, Terminalia is divided into two groups: an African group with a few taxa from Asia and the Pacific islands and an Asian group. Our results could be used for good reference about the classification of the genus Terminalia.
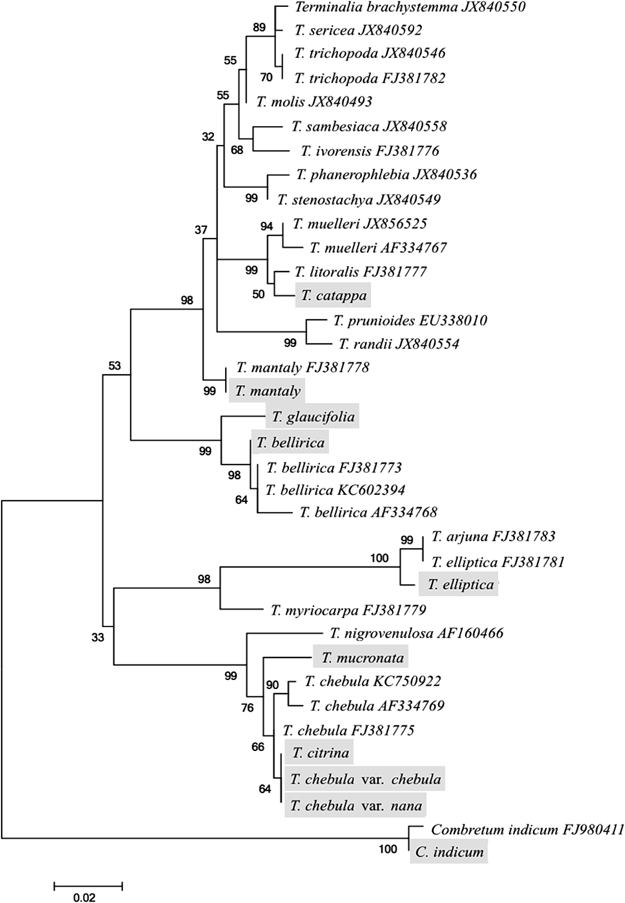
Based on approximately 0.6 kb aligned nucleotide sequence of nuclear ITS1-5.8S-ITS2 region. Numbers at nodes indicate bootstrap values with 1000 replications. Branch lengths are proportional to the number of substitutions per site (refer to scale bar). Sequence data of species in gray brackets were obtained in the present study and those of other species were retrieved from the DNA database.
Vast improvements in the identification and analysis of SNPs in plants were noted in last ten years. Such methods as PCR-RFLP and ARMS, which use PCR markers based on SNPs, have been adopted for the authentication of herbs.
Of the Terminalia samples collected in Thailand, five had medicinal value, including T. chebula var. chebula, T. chebula var. nana, T. citrina, T. bellirica, and T. catappa. We subjected T. chebula var. chebula, T. bellirica, and T. catappa to PCR-RFLP analysis. Because, examination of the aligned nucleotide sequences in the ITS1-5.8S-ITS2 regions revealed almost identical nucleotide sequences in T. chebula var. chebula, T. chebula var. nana, and T. citrina. Therefore, the PCR-RFLP method could not differentiate them. The amplified fragments of these three Terminalia species and P. emblica were approximately 800 bp long. The nucleotide sequences showed species-specific sequences at sites subjected to the restriction enzyme analysis. To exclude T. catappa, the XspI restriction enzyme was used for diagnosis. Three fragments distinct to T. catappa were found. XspI recognized 5′-CTAG-3′, which was found in the ITS region of T. catappa only at two sites, whereas T. chebula var. chebula, T. bellirica, and P. emblica could not to be digested at the same sites. XspI digestion cleaved the nucleotide sequence of T. catappa was cleaved into three amplicons that were approximately 200, 408, and 200 bp long (Fig. 1A). The cleaved products of T. catappa appeared as two bands in 3.0% Tris–acetate–ethylenediaminetetraacetic acid (TAE) agarose gel electrophoretogram (Fig. 3A). Then, to discriminate T. chebula var. chebula, T. bellirica, and P. emblica, the Aor13HI restriction enzyme was used to recognize the specific nucleotides in the three species. It was found that Aor13HI recognized 5ʹ-TCC GGA-3ʹ. As shown in Fig. 3B, the amplified sequence of T. bellirica was cleaved into two fragments that were approximately 550 and 250 bp long, and that of T. chebula var. chebula was cleaved into three fragments that were approximately 200, 350, and 250 bp long, P. emblica could not to be digested by the same restriction enzyme.

Three percent TAE agarose gel electrophoregram of PCR product of P. emblica, T. bellirica, T. catappa, and T. chebula var. chebula generated by primers Ter.Af and Ake-26SR, and then digested with restriction enzyme. A. XspI (C^TAG). The ITS1-5.8S-ITS2 fragments of P. emblica, T. bellirica, T. catappa, and T. chebula var. chebula were amplified, and digested fragments (left 4 lanes) and non-digested fragments (right 4 lanes) were separated by agarose gel electrophoresis, lane M, 1 kb DNA ladder (BioTools Inc., Japan). B. Aor13HI (T^CCGGA). The amplified fragments of P. emblica, T. bellirica, and T. chebula var. chebula in the ITS1-5.8S-ITS2 region were separated into the cleaved amplicons (left 3 lanes), lane M, and uncleaved amplicons (right 3 lanes).
SNPs were noted in the nucleotide sequences of the ITS regions of medicinal Terminalia species (Table 4). Different nucleotides at position nos. 93–94, 215–217, and 267–277 were used to design species-specific primers for the authentication of T. catappa, T. chebula var. chebula, and T. bellirica, respectively. The reverse primers of the three diagnostic primer pairs had the same sequences (Table 3). Therefore, three specific primers TCA.Af/Ter.Br, TCH.Af/Ter.Br, and TBE.Cf/Ter.Br, were fabricated for discriminating T. catappa, T. chebula var. chebula, and T. bellirica, respectively. The amplified products from those primer sets had different sizes: T. catappa, 388 bp; T. chebula var. chebula, 266 bp; and T. bellirica, 209 bp (Fig. 4). As a consequence, the three pairs of diagnostic primers could be used to authenticate the medicinal Terminalia plants.

Lanes 1–4 are the same as species-specific primer pairs: 1. PHE.Bf/PHE.Br, 2. TCA.Af/Ter.Br, 3. TCH.Af/Ter.Br, and 4. TBE.Cf/Ter.Br. Lane M is 1 kb DNA ladder.
The two methods for separating the three medicinal Terminalia species and P. emblica, PCR-RFLP and ARMS, were used to authenticate Terminalia crude drugs including P. emblica crude drugs and the ingredients of Triphala, which consisted of T. chebula, T. bellirica, and P. emblica (or Samo Thai, Samo Phiphek, and Makampom, respectively, the Thai names of their crude drugs) in the ratio of 1 : 1 : 1. The PCR-RFLP method could be developed for the authentication of the three medicinal Terminalia species, but not P. emblica. The ARMS method, on the other hand, could be applied also for P. emblica with simplicity and efficiency.24) The nucleotide sequence of the ITS region of P. emblica was aligned and searched for different sites for designing the specific primer. The result showed that amplicon of 497 bp was amplified by PHE.Bf/PHE.Br only for P. emblica (Fig. 4), which can authenticate its crude drug. The amplification was conducted with the combination of these species-specific primers under identical concentration and temperature conditions. The multiplex-ARMS-PCR amplification efficiently produced three apparently amplified PCR products as shown in Fig. 5. Therefore, this technique enabled the authentication of Terminalia crude drugs and the ingredients of Triphala. For instance, the 209 bp fragment was found only in the genomic DNA sample of Samo Phiphek. Three similar fragments that were 209, 266, and 497 bp long were found in the genomic DNA samples of Triphala. These results clearly showed that ARMS is effective for the identification of Terminalia crude drugs and Triphala formulations.

The authors are grateful to Wannaree Charoensup (Botanist, Faculty of Pharmacy, Chiang Mai University, Thailand), and Drs. Ratchuporn Suksathan and Monthon Norsaengsri (Queen Sirikit Botanic Garden, Thailand) for their kind assistance in preparing and identifying Terminalia specimens.
The authors declare no conflict of interest.