2018 年 41 巻 3 号 p. 338-341
2018 年 41 巻 3 号 p. 338-341
Regulating γ-aminobutyric acid (GABA) uptake transport on the plasma membranes is required for its efficient clearance from the brain interstitial fluid. The purpose of this study was to clarify the assembly of taurine transporter (TauT/Slc6a6) and PSD-95/Disc-large/Zo-1 (PDZ) domain of Na+–H+ exchanger regulatory factor 1 (NHERF1) as a regulatory mechanism of TauT-mediated GABA transport activity. In vitro glutathione S-transferase (GST)-pull down assay and immunoblotting with anti-NHERF1 antibody revealed that NHERF1 protein was present in rat brain lysates as the binding protein of the GST-fusion TauT C-terminal protein with the PDZ-binding ETMM motif but not its corresponding deletion mutant lacking the motif. [3H]GABA uptake by TauT-NHERF1-coexpressing oocytes and TauT-singly expressing oocytes exhibited saturable kinetics with Michaelis–Menten constant values of 0.835±0.288 and 0.982±0.569 mM and a maximal transport velocity of 206±37 and 283±28 pmol/(h·oocyte), respectively. These results suggest that the assembly of TauT PDZ-binding motif and NHERF1 increases the maximal transport velocity of GABA rather than changes the affinity.
Regulating the localization and function of neurotransmitter uptake transporters on the plasma membranes is required for the efficient clearance of neurotransmitters from the synaptic cleft in the brain. Previous reports have demonstrated that the astrocyte uptake of glutamate, an excitatory neurotransmitter, is increased by the scaffolding interactions between the C-terminus of glutamate transporter (GLAST/Slc1a3) protein and the PSD-95/Disc-large/Zo-1 (PDZ) domain of Na+–H+ exchanger regulatory factor 1 (NHERF1), a scaffolding protein.1) This report supports the belief that assembly of neurotransmitter transporters and NHERF1 is a regulatory mechanism of neurotransmitter transport in the brain.
Our previous results have demonstrated that rat taurine transporter (TauT/Slc6a6) accepts γ-aminobutyric acid (GABA), a suppressive neurotransmitter, as a low-affinity substrate with a Michaelis–Menten constant (Km) value of 1.5 mM.2) This raises the possibility that TauT is a clearance system of GABA in the brain interstitial fluid (ISF), which may compensate for the high-affinity GABA transporters-mediated GABA uptake.3) It has been reported that TauT is expressed in the astrocytes4) as is the case in NHERF1.1) Furthermore, the PDZ-binding motif (ETMM) at the C-terminus of rat TauT exhibits a similar consensus sequence of GLAST (ETKM) which has been reported to be associated with the PDZ domain of NHERF1.1) These lines of evidence prompted us to hypothesize that NHERF1 is a regulatory factor of TauT-mediated GABA transport activity on the plasma membranes. Accordingly, the purpose of this study was to clarify the interaction between NHERF1 and the TauT C-terminus and its impact on TauT-mediated GABA transport activity.
4-Amino-n-[2,3-3H]butyric acid ([3H]GABA, 87.0 Ci/mmol) was obtained from GE Healthcare (Chalfont St. Giles, U.K.). All other chemicals were commercial products of analytical grade. Male Wistar rats (7-weeks old) were purchased from Japan SLC, Inc. (Shizuoka, Japan). The rats were maintained on a 12-h light/dark cycle in a temperature-controlled environment with free access to food and water. Mature female Xenopus laevis frogs were obtained from Kato-S-Science (Chiba, Japan). All experiments were approved by the Animal Care Committee at the University of Toyama and Tohoku University.
Glutathione S-Transferase (GST) Pull-Down AssayThe whole brain tissues were homogenized in the buffer A [20 mM N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (HEPES)–Tris (pH 7.4), 120 mM NaCl, 0.6% Triton X-100, 5 mM ethylenediaminetetraacetic acid (EDTA), and a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany)], and incubated for 2 h at 4°C with gentle rotation. The homogenates were centrifuged at 27000×g for 30 min at 4°C and the supernatant was collected. Protein concentrations were determined using the DC protein assay reagent (Bio-Rad, Hercules, CA, U.S.A.). The cDNAs encoding the polypeptides of the TauT C-terminus and its deletion mutant (Table 1) were amplified by RT-PCR using the primers as shown in Table 2 and inserted in the pGEX4T-2 plasmid vector (GE Healthcare). The polypeptides were expressed as GST fusion proteins in Escherichia coli (BL21-codon plus RIL-competent cells; Stratagene, La Jolla, CA, U.S.A.). The fusion protein was purified with glutathione-sepharose 4B beads (GE Healthcare) according to the manufacturer’s instructions. The GST fusion protein beads (10 µg) in the buffer A were then incubated with the brain lysate (4 mg) at 4°C overnight. The beads were washed with the buffer A and re-suspended in sodium dodecyl sulfate (SDS) sample buffer. A protein sample was separated by SDS-polyacrylamide gel electrophoresis (PAGE), and bands were electrotransferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, CA, U.S.A.). The blotted membranes were incubated with rabbit anti-rat NHERF1 antibody (1 µg/mL; Sigma, St. Louis, MI, U.S.A.) overnight at 4°C in the Block Ace (Dainippon-Pharm, Osaka, Japan), and incubated for 1 h at 25°C with horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA, U.S.A.), and visualized with an enhanced chemiluminescence kit (GE Healthcare).
| Name | Amino acid sequence |
|---|---|
| TauT-CT1 | ITPREPNRWAVEREGATPFHSRATLMNGALMKPSHVIVETMM |
| TauT-CT1Δ3 | ITPREPNRWAVEREGATPFHSRATLMNGALMKPSHVIVE– – – |
| TauT-CT2 | REGATPFHSRATLMNGALMKPSHVIVETMM |
| TauT-CT2Δ3 | REGATPFHSRATLMNGALMKPSHVIVE– – – |
| Upstream primer (5′ to 3′) | Downstream primer (5′ to 3′) | Annealing temperature (°C) | |
|---|---|---|---|
| TauT-C1 | ggatccataacccccagggagcccaa | gaattctcacatcatggtctccacaatg | 60 |
| TauT-C1Δ3 | ggatccataacccccagggagcccaa | gaattctcactccacaatgacgtgactggg | 60 |
| TauT-C2 | ggatcccgtgaaggggctacgcccttt | gaattctcacatcatggtctccacaatg | 60 |
| TauT-C2Δ3 | ggatcccgtgaaggggctacgcccttt | gaattctcactccacaatgacgtgactggg | 60 |
The underline represents the recognizing sequence of restricting enzymes of EcoRI (ggatcc) and BamHI (gaattc).
Capped cRNA was transcribed using T7 RNA polymerase from NotI-linearized pGEM-HEN containing an open reading frame of rat TauT5) (GenBank accession number NM_017206) and NHERF1 (GenBank accession number NM_021594) cDNAs. The capped cRNA (30–50 ng) dissolved in water (23 nL) or water (23 nL) was injected in defolliculated Xenopus laevis oocytes. The oocytes were then incubated at 18°C in freshly prepared standard oocyte saline (SOS) solution (100 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES, 25 µg/mL gentamycin, 2.5 mM pyruvate and 1% bovine serum albumin, pH 7.5). After incubation for 4 to 6 d, experiments were performed. For the detection of NHERF1 protein, the oocytes were homogenized with the SDS sample buffer and immunoblotted with anti-NHERF1 antibody as described above. The uptake of [3H]GABA (14 nM) by the oocytes was estimated according to our previously reported method.6) The uptake amount of [3H]GABA in the oocytes was expressed as the oocyte-to-medium (oocyte/medium) ratio (µL/oocyte), which was calculated as the amount of [3H]GABA in oocytes (dpm/oocyte) divided by the concentration of [3H]GABA in the medium (dpm/µL).
Kinetic AnalysesThe kinetic parameters for GABA uptake by Xenopus laevis oocytes were determined from the following equation:
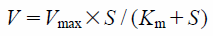 |
Except for kinetic parameters, all data are shown as the mean±standard error of the mean (S.E.M.) while kinetic parameters are presented as the mean±standard deviation (S.D.). The significance of differences between two group means was determined by an unpaired, two-tailed Student’s t-test. The statistical significance of differences among means of more than two groups was assessed by one-way ANOVA followed by the modified Fisher’s least-squares difference method.
To determine whether NHERF1 has the ability to bind the TauT C-terminal with the ETMM motif, an in vitro GST-pull down assay was performed. Two sets of GST-fusion proteins with different lengths were generated in the E. coli: the wild-type TauT C-terminus with the ETMM motif (GST-TauT-CT1 and GST-TauT-CT2) and its corresponding deletion mutant without the ETMM motif (GST-TauT-CT1Δ3 and GST-TauT-CT2Δ3). The rat brain lysate was incubated with the individual GST-fusion proteins and the complex of the GST-fusion protein and its binding proteins were collected by glutathione-sepharose 4B beads and resolved by SDS-PAGE. Immunoblotting with anti-NHERF1 antibody was performed to clarify whether NHERF1 was detected as the TauT C-terminus binding protein. The bands as indicated by arrowheads were detected as the binding proteins with wild-type TauT C-terminus polypeptides with a size of 55 kDa, which corresponded to that of NHERF1 protein8) (Figs. 1A, B; TauT-CT1 and TauT-CT2, respectively). The molecular size of the bands as indicated by the arrowheads was consistent with those detected in the brain homogenates (Figs. 1A, B; Brain lysate). In contrast, no band was detected in the binding proteins with TauT C-terminus deletion mutant polypeptides (Figs. 1A, B; TauT-CT1Δ3 and TauT-CT2Δ3, respectively). These results indicated that the TauT C-terminus with the ETMM motif could be bound to NHERF1.

In vitro GST-pull down assay. GST fusion proteins containing the TauT C-terminus [GST–TauT-CT1 and GST–TauT-CT2] and the TauT C-terminus deletion mutant [GST–TauT-CT1Δ3 and GST–TauT-CT2Δ3] were immobilized on glutathione–sepharose beads and incubated with total brain lysate. Bound proteins with GST–TauT-CT1 (A), GST–TauT-CT1Δ3 (A), GST–TauT-CT2 (B), and GST–TauT-CT2Δ3 (B) or the brain lysate (A, B) used as the positive control were resolved on SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-NHERF1 antibody.
As shown in Fig. 2, a single band corresponding to NHERF1 was detected in the TauT- and NHERF1-coexpressing Xenopus oocytes (TauT-NHERF1/oocytes) at the corresponding size of the NHERF1 protein. No band corresponding to NHERF1 was detected in the water-injected oocytes and TauT-expressing Xenopus oocytes (TauT/oocytes). As shown in Fig. 3, the uptake activity of [3H]GABA was measured in TauT-NHERF1/oocytes, TauT/oocytes, NHERF1-expressing Xenopus oocytes (NHERF1/oocytes), and water-injected oocytes. [3H]GABA uptake by TauT/oocytes and TauT-NHERF1/oocytes was 2.5- to 3.7-fold greater than that by water-injected oocytes and NHERF1/oocytes. These results indicated that TauT protein was abundantly expressed in TauT/oocytes and TauT-NHERF1/oocytes. The uptake by TauT-NHERF1/oocytes was 1.5-fold greater than that by TauT/oocytes and this was statistically significant. As shown in Fig. 4, the [3H]GABA uptake by TauT-NHERF1/oocytes and TauT/oocytes exhibited saturable kinetics with Km values of 0.835±0.288 and 0.982±0.569 mM and Vmax values of 206±37 and 283±28 pmol/(h·oocyte), respectively. The statistical significance of the Km and Vmax values between TauT-NHERF1/oocytes and TauT/oocytes was estimated to be p=0.818 and p=0.094, respectively. Although the initial uptake rate at 6 mM was reduced compared with that at 4 mM in TauT-NHERF1/oocytes and TauT/oocyte, the initial uptake rates at 4 and 6 mM in TauT-NHERF1/oocytes were greater than those in TauT/oocytes. These results indicated that the co-expression of TauT and NHERF1 tended to increase the maximal GABA transport activity (Vmax) rather than change the affinity (Km).


Each column represents the mean±S.E.M. (n=10). * p<0.01, significant difference between the indicated groups.

The inset graph shows the Eadie–Scatchard plot of the same data. Each point represents the mean±S.E.M. (n=10).
The present study has demonstrated that assembly of TauT PDZ-binding motif and NHERF1 tends to increase the maximal transport velocity (Vmax) of GABA rather than change the affinity (Km). This finding is in good agreement with the previous result showing that the association of organic anion transporter (OAT) 4 (SLC22A11) and NHERF1 increases the Vmax of estron-3-sulfate without changing the Km value.9) The mechanism has been explained by the increased surface expression of OAT4 protein. Several reports have also demonstrated that the assembly of NHERF1 and the PDZ domain of transporters such as GLAST,10) organic anion transporter OATP1A2,11) and phosphate transporter NPT2a12) increases the plasma membrane localization of the transporters. It has also been reported that the association of PDZK1 and organic cation/carnitine transporter OCTN2 (SLC22A5) increases the Vmax of carnitine whereas the cell-surface expression level of OCTN2 is not significantly increased.13) This previous result suggests that PDZK1 is a functional regulator of OCTN2 through direct interaction with the C-terminus of OCTN2. Based on the present findings and the previous reports, it is likely that NHERF1 has the potential for stabilizing/anchoring TauT protein at the plasma membrane or acting as a functional regulator of TauT-mediated GABA transport.
The increase in TauT-mediated GABA uptake by the TauT-NHERF1 assembly may be of pathological significance under ischemic conditions. It has been reported that the GABA concentration during cerebral ischemia in human brain interstitial fluid is increased to a maximum of 35.7 µM which is approximately 600-fold greater than the normal concentration (0.06 µM).14) Since the Km or half maximal IC50 values of GABA transporters (Slc6a1, Slc6a11, and Slc6a13)-mediated GABA transport in the brain has been estimated to be in the order of 1–30 µM3,15) in rodents, the GABA clearance activity of GATs may be over 50% saturated under ischemic conditions. It has also been reported that the astrocyte expression level of GAT3/Slc6a11 is reduced in transient focal ischemia in rats.16) Therefore, it seems plausible that TauT-mediated GABA uptake with a low affinity and high capacity would contribute to the regulation of GABA level in the synaptic cleft by increasing the TauT-NHERF1 assembly.
In conclusion, NHERF1 plays a role in increasing TauT-mediated GABA transport by assembly with the PDZ-binding motif of TauT.
We thank Dr. T. Abe for supplying the pGEM-HEN vector. This study was supported, in part, by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) for Scientific Research on Innovative Areas [KAKENHI: 16H01326].
The authors declare no conflict of interest.