2018 年 41 巻 4 号 p. 536-545
2018 年 41 巻 4 号 p. 536-545
Lactobacillus plantarum SN35N, which has been previously isolated from pear, secretes exopolysaccharide (EPS). The aim of the present study is to characterize the EPS chemically and to find the EPS-biosynthesizing gene cluster. The present study demonstrates that the strain produces an acidic EPS carrying phosphate residue, which is composed of glucose, galactose, and mannose at a molecular ratio of 15.0 : 5.7 : 1.0. We also show that acidic EPS strongly inhibits the catalytic activity of hyaluronidase (EC 3.2.1.35), promoting an inflammatory reaction. In the present study, we also determined the complete genome sequence of the SN35N strain, demonstrating that the genome is a circular DNA with 3267626 bp, and the number of predicted coding genes is 3146, with a GC content of 44.51%. In addition, the strain harbors four plasmids, designated pSN35N-1, -2, -3, and -4. Although four EPS-biosynthesizing genes, designated lpe1, lpe2, lpe3, and lpe4, are present in the SN35N chromosomal DNA, another EPS gene cluster, lpe5, is located in the pSN35N-3 plasmid, composed of 35425 bp. EPS low-producing mutants, which were obtained by treating SN35N cells with novobiocin, lost the lpe5 gene cluster in the plasmid-curing experiment, suggesting that the gene cluster for the biosynthesis of acidic EPS is present in the plasmid. The present study shows the chemical characterization of the acidic EPS and its inhibitory effect to the hyaluronidase.
Probiotics are defined as “living microorganisms conferring a health benefit to the host, when administered in adequate amounts.”1) Lactic acid bacterium (LAB), some strains of which are known as a probiotic, is a generic name given to non-pathogenic Gram-positive bacteria that produce one or two moles of lactic acid from one mole of sugars during fermentation. LABs, which are also generally recognized as safe (GRAS) microorganisms, have been traditionally used to make fermented foods such as fermented dishes, yogurt, and cheese.2–4)
Some LAB strains give health benefits to human, such as immunomodulation and improvement of intestinal disorders. It has been also reported that an LAB strain has a potent effect on preventing and improving obesity, and helps to decrease serum lipids and cholesterol.5–8) Until now, we have established a plant-derived LAB library which consist of more than 600 strains from medicinal plants, vegetables, flowers, and fruits.9–11) We have searched LAB strains useful for preventive medicine and found several strains that enhance intestinal immunity, improve liver function, and prevent metabolic syndrome.12–15)
Exopolysaccharide (EPS) produced by some LAB strains16,17) exhibits immunomodulation, anti-gastritis and anti-ulcer functions, and anti-virus activities.18–20) Although LAB strains numbered SN13T and SN35N13) isolated previously by our group were identified as Lactobacillus (Lb.) plantarum, only the SN35N strain produces EPS. We have also isolated two EPS-producing Pediococcus (P.) pentosaceus, named LP28 and LY45 strains. We have found using high-fat diet-induced obese mice that the intake of the LP28 strain effective for reducing body weight gain, improving fatty liver, and decreasing accumulated abdominal visceral fat.15) In addition, we have previously determined the strain-specific EPS biosynthetic gene cluster in EPS-producing P. pentosaceus LP28.21) On the other hand, the EPS produced by LY45 strain has been found to inhibit the enzymatic activity of hyaluronidase.22) Hyaluronic acid, which is generated by digestion with hyaluronidase, stimulates the inflammatory response reaction.23,24) Therefore, a compound (substance) that inhibits hyaluronidase may become a candidate for an anti-inflammatory agent.
In the present study, we have analyzed the monosaccharide components and the acidic residue of the acidic EPS produced by a pear-derived Lb. plantarum SN35N. Additionally, we evaluate partially the healthcare function, demonstrating that the SN35N-derived acidic EPS strongly inhibits the catalytic activity of hyaluronidase promoting inflammatory reactions. Furthermore, we have found EPS-biosynthesizing gene clusters on the SN35N chromosome and on a plasmid by sequencing the whole genome together with the plasmid DNA. To generate a mutant that scarcely produces EPS, we treated the SN35N cells with novobiocin, demonstrating that the pSN35N-3-cured mutant did not produce the acidic EPS. A gene cluster necessary for the biosynthesis of the acidic EPS may be present on the plasmid.
De Man, Rogosa, and Sharpe (MRS) broth (Merck KGaA, Darmstadt, Germany) was used as a culture medium for Lb. plantarum SN35N. A semi-defined medium (SDM)25) supplemented with a 0.2% (v/v) vitamin solution and a 0.1% (v/v) trace element solution26) instead of a yeast nitrogen base, called modified SDM, was used to produce the EPS.
Culture Condition for Producing EPSFor the seed culture, a portion of the SN35N cells frozen stock solution was inoculated into fresh MRS broth and grown at 37°C until the stationary phase of growth under the condition of standing culture. The seed culture was inoculated at 0.5% (v/v) into a modified SDM medium and incubated at 28°C for 2 d under the stand culture condition without shaking.
Purification of EPS from the SN35N Culture BrothEPS was purified from the culture broth in accordance with the method described previously22): After the addition of trichloroacetic acid (TCA) to the SN35N culture broth, the LAB cell mass and proteins were removed from the cultured broth by centrifugation. The resulting supernatant fluid was mixed with acetone to precipitate the EPS. The nucleotides and proteins in the precipitated EPS were digested with deoxyribonuclease I (Worthington Biochemical Corporation, Lakewood, NJ, U.S.A.), ribonuclease A (Nacalai Tesque, Kyoto, Japan), and proteinase K (Wako Pure Chemical Industries, Ltd., Osaka, Japan). After adding the TCA, the protein and debris were removed by centrifugation, and the crude EPS was obtained from the resulting supernatant fluid by ethanol precipitation. The resulting EPS pellet was dissolved into distilled water. Prior to determination of the EPS content by the phenol sulfate method,27) the crude EPS solution was dialyzed against the distilled water using an Amicon Ultra (MWCO=10 kDa, Merck Millipore Ltd., Carrigtwohill, Co., Cork, Ireland).
The acidic EPS was purified from the crude EPS by using a TOYOPEARL DEAE-650M column (2.5×22 cm; Tosoh Bioscience, Tokyo, Japan) with the method described previously.22) The acidic EPS was eluted from the column with a linear gradient of NaCl (0 to 240 min, 0 mM; 240 to 600 min, 0–500 mM). The EPS-contained fractions were pooled and dialyzed against the distilled water by using an Amicon Ultra (MWCO=10 kDa).
Hyaluronidase Inhibitory AssayThe assay for hyaluronidase inhibition was performed according to the protocol established by Fujitani et al.,28) with a slight modification method.22) The inhibitory activity was compared by calculating the IC50 value, which is defined as the EPS concentration inhibiting 50% enzyme activity.
Acute Toxicity Using Rats and Mutagenicity TestsAn acute oral toxicity experiment of the SN35N cells, which was orally administrated, was done through the New Drug Development Research Center, Inc. (the protocol numbers are 06060-1 and 06060-2). The experiment was performed according to the Principles of Good Laboratory Practice. Five-week-old Crl: Ceasarean Derived (Sprague–Dawley) male rats were purchased from Charles River Laboratories Japan, Inc. LAB cells were resuspended into the purified water (Yakuhan Pharmaceutical Co., Ltd., Hokkaido, Japan). The rats were divided into three groups of five rats each and housed in stainless steel cages under controlled temperature (22±3°C) and 12 h light-dark cycles. Rats had free access to CRF-1 diets (Oriental Yeast Co., Ltd., Tokyo, Japan) and water. After 1 week of acclimation, each group was assigned an LAB-fed group and a reference group: a high-dose group (1.5×1012 colony-forming unit (CFU)/kg), a low-dose group (0.75×1012 CFU/kg), and a pure water group. Cell suspensions or purified water was orally administrated to rats by using a sterile probe once a day for 2 weeks. During the experiment, rats’ exercise activity, behavior, general health status, and body weight were recorded at 1-, 3-, 7-, 10-, and 14-d points. After the experimental period, the rats were euthanized, and histological analyses of some extracted organs were performed. The same experiment was also carried out using female rats.
The mutagenicity test (umu test) of the Lb. plantarum SN35N culture broth was performed using an Umulac AT-F kit (Protein Purify Co., Ltd., Gunma, Japan) in accordance with the manufacturer’s instructions.
Molecular Mass AnalysisThe molecular mass of the acidic EPS was estimated using an HPLC system equipped with gel-filtration chromatography with a SUGAR KS-806 column (Showa Denko, Tokyo, Japan). The analytical conditions were as follows: ultrapure water was used as a mobile phase at a flow rate of 0.7 mL/min. The column oven temperature was set at 80°C, and the eluent was monitored by the RI detector. The molecular mass was calculated from the calibration curve made using pullulan standards.
Monosaccharide Composition of EPSThe composition of monosaccharide in the acidic EPS was analyzed after the hydrolysis reaction as follows: the purified EPS was dissolved in 10 mL of purified water. After adding 300 µL of 18 M H2SO4, the samples were hydrolyzed for 3.5 h at 110°C. The hydrolysate was neutralized by adding Ba(OH2)·8H2O; it was then filtrated with a 0.45 µm pore-sized membrane filter. The resulting filtrate was applied on an HPLC system equipped with a SUGAR SP0810 column (Showa Denko). The analytical conditions were as follows: ultrapure water was used as a mobile phase at a flow rate of 0.7 mL/min. The analysis was performed at 80°C, and the eluates from the column were monitored with an RI detector.
Detection of Phosphate ResidueBIOMOL Green Reagent (Biomol GmbH, Hamburg, Germany) was used to analyze the phosphate residue in acidic EPS. The EPS was dissolved into distilled water at a final concentration of 0.02% (w/v). Five milliliter of EPS solution mixed with 1 mL of 40 g/L potassium peroxodisulfate was incubated at 120°C for 30 min, and then cooled to room temperature (r.t.). A sample not subjected to heat treatment was used as a control. In this case, distilled water was used instead of the EPS solution as a blank test sample.
One milliliter of BIOMOL Green Reagent and a 100 µL aliquot of 4-times diluted measuring samples were mixed and reacted at r.t. for 25 min. The phosphate release was confirmed by measuring A620. The quantity of phosphate was calculated form the standard curve prepared by using serial dilutions of phosphate with 5–80 µM.
DNA PreparationThe SN35N chromosomal DNA was isolated as described previously,10) with a slight modification: the cell mass was harvested from the culture broth by centrifugation and washed with a glucose–ethylenediaminetetraacetic acid (EDTA) buffer (50 mM glucose, 10 mM EDTA, 25 mM Tris–HCl, pH 8.0). The washed cells were resuspended into the same buffer, containing 10 µg/mL ribonuclease A (Nacalai Tesque), 4 mg/mL lysozyme (Wako Pure Chemical Industries, Ltd.), and 0.4 mg/mL achromopeptidase (Wako Pure Chemical Industries, Ltd.). After incubation for 3 h at 37°C, the cells were lysed by adding 0.1 volume of 10% (w/v) sodium dodecyl sulfate, and the proteins were denatured and removed by chloroform/isoamyl alcohol extraction. Finally, the chromosomal DNA was purified by ethanol precipitation.
Genome Sequencing and AnnotationThe whole genome of the SN35N strain was sequenced on a next-generation sequencing platform, PacBio RS II (Pacific Biosciences, Menlo Park, CA, U.S.A.), on a single molecule real-time (SMRT) cell using P6 polymerase and C4 chemistry (P6C4); the purified genomic DNA was fragmented using a g-TUBE (Covaris, Woburn, MA, U.S.A.), and the sheared short fragments were then purified using an AMPure PB kit (Pacific Biosciences). A PacBio DNA Template Prep Kit 1.0 (Pacific Biosciences) and a PacBio DNA/Polymerase Binding Kit P6 (Pacific Biosciences) were used for constructing the DNA library. The inadequate short fragments were eliminated by Blue Pippin (Sage Science, Beverly, MA, U.S.A.), and the resulting purified DNA library was then sequenced on the PacBio SMRT platform. The hierarchical genome assembly process (HGAP) protocol29) was used for de novo assembling, and the resulting genome contig was annotated by the Microbial Genome Annotation Pipeline (MiGAP). The genome sequence was analyzed using in silico Molecular Cloning Genomics Edition (In Silico Biology, Inc., Kanagawa, Japan). The GenBank/EMBL/DDBJ accession numbers for the sequences reported in the present study are AP018405 (for chromosomal DNA), AP018406, AP018407, AP018408, and AP018409 (for plasmid DNAs).
Generation of an EPS Non-producing MutantA mutant from Lb. plantarum SN35N, which cannot produce EPS, was isolated by treatment with novobiocin; an aliquot of the overnight culture was inoculated into MRS broth supplemented with 0.8 µg/mL novobiocin, useful as a curing agent for plasmid. After cultivation for 2 d, a portion of the culture was plated on fresh MRS agar and incubated anaerobically for 2 d until colonies appeared. The existence of an lpe5 cluster in each colony was confirmed by PCR analysis, using sense (5′-catcgcaattatgatcaagcgcg-3′) and anti-sense (5′-gacgctgcttagcatactcacta-3′) primers. The EPS productivity by the plasmid-cured (EPS non-producing) mutant, which was named (SN35N-Δp3), was also evaluated by the same method as for SN35N parental strain.
Lb. plantarum SN35N grown in a modified-SDM medium produces EPS outside the cells. The chromatography profile when using a TOYOPEARL DEAE-650M anion-exchange column shows that the SN35N-derived EPS displays an acidic property. The yield of acidic EPS from the culture broth was estimated to be 48.2 mg/L (Fig. 1). The molecular mass, as estimated by a gel-filtration HPLC profile, was approximately 250 kDa.
As shown in Fig. 2 and Table 1, the HPLC profile shows that the acidic EPS is composed of glucose, galactose, and mannose at a monosaccharide ratio of 15.0 : 5.7 : 1.0, respectively. Thus, mannose is slightly detectable in the SN35N-derived EPS. Monosaccharides mainly present in EPS produced by other LAB strains are glucose and galactose.32) Using phosphate quantification analysis of acid-hydrolyzed acidic EPS, we show that the acidic EPS from the SN35N strain contains 1.0 µmol phosphorylate residue per 1 mg of EPS.

Dashed lines indicate the NaCl gradient concentrations in the eluates. The presence of EPS was monitored at 490 nm by the phenol sulfate method (solid line).
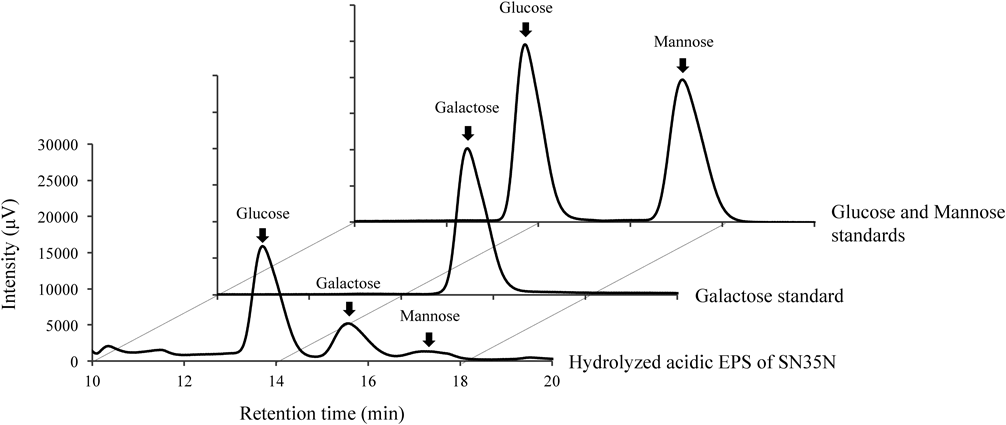
Each monosaccharide peak was identified using the monosaccharide standard solution.
| Saccharide | r.t. (min) | Relative molar ratio |
|---|---|---|
| Glucose | 13.70 | 15.0 |
| Galactose | 15.55 | 5.7 |
| Mannose | 17.21 | 1.0 |
We carried out a kinetic analysis to evaluate hyaluronidase’s inhibitory effect on the SN35N-derived acidic EPS. The IC50 value (240 µg/mL) was lower than that of fucoidan (from Laminaria Japonica, 2000<µg/mL). On the other hand, that of the P. pentosaceus LY45-derived acidic EPS, which was previously isolated by our group, was 1300 µg/mL.22) The IC50 values of sodium cromoglicate and dipotassium glycyrrhizinate, which are well-known as anti-inflammatory agents, were 100 and 530 µg/mL, respectively.22)
Safety Evaluation of SN35N StrainAcute oral toxicity tests for the SN35N cells demonstrated that significant activity changes and feeding-related illness or death of rats were not observed. Obvious differences and inflammatory symptoms were also not observed in some organs in the rats. In addition, a mutagenicity test for the SN35N culture broth did not give rise to mutagenicity.
Genome Sequence of the SN35N StrainWe determined the whole-genome sequence of the SN35N strain. The sequence information indicates that the whole genome is a circular DNA, and the size is 3267626 bp, with a GC content of 44.51%. The number of CDS (coding sequence) is predicted to be 3146 (Table 2), when compared with other eight Lb. plantarum strains deposited in the DDBJ database, in addition to the data published previously.30,31,33–37) The average genome size of several Lb. plantarum strains is 3286 kb, with 44.36% of GC content and 3057 CDS, showing that the values of other Lb. plantarum genomes are almost the same as that of the SN35N genome.
| Strain | Genome size (bp) | GC content (%) | CDS | tRNA genes | rRNA genes | Isolation origin | Accession no. | Reference |
|---|---|---|---|---|---|---|---|---|
| SN35N | 3267626 | 44.5 | 3146 | 75 | 16 | Pear | AP018405 | This study |
| WCFS1 | 3308273 | 44.5 | 3013 | 70 | 15 | Human saliva | NC_004567 | 34 |
| JDM1 | 3197759 | 44.7 | 2904 | 61 | 16 | Human intestinal tract | NC_012984 | 33 |
| ZJ316 | 3203964 | 44.4 | 2894 | 61 | 15 | Healthy newborn fecal sample | NC_020229 | 30 |
| 16 | 3044678 | 44.7 | 2784 | 66 | 16 | Malt production steep water | NC_021514 | 31 |
| B21 | 3284260 | 44.5 | 3021 | 63 | 17 | Vietnamese fermented sausage (nemchua) | NZ_CP010528 | 36 |
| HFC8 | 3067675 | 44.3 | 2766 | 68 | 16 | Faecal sample | NZ_CP012650 | 35 |
| KP | 3418468 | 44.3 | 3184 | 81 | 16 | Whole fly | NZ_CP013749 | 37 |
| DF | 3423963 | 44.4 | 3204 | 81 | 16 | Whole fly | NZ_CP013753 | 37 |
The predicted EPS-biosynthesizing gene clusters, designated lpe1, pe2, lpe3, and lpe4, are present in the chromosomal DNA (Table 3 and Fig. 3). Lpe1, which is smallest of the four clusters, contains only four open reading frames (ORFs) predicted to be involved in EPS biosynthesis (lpe1A, lpe1B, lpe1C, and lpe1D). The lpe2 and lpe3 clusters, which are found on a region adjacent to lpe1, are composed of 10 (lpe2A–J) and 11 (lpe3A–K) ORFs, respectively. The lpe4 cluster is 880 kb away from the lpe1–3 region. Among these clusters, only the lpe2 and lpe4 seem to include genes necessary for the biosynthesis of EPS—the priming glycosyltransferase (lpe2E and lpe4E), glycosyltransferase (lpe2F, lpe2H, lpe2I, lpe4F, lpe4G, and lpe4I), flippase (lpe2A, lpe2J, and lpe4J), polymerase (lpe2G and lpe4H), and chain length regulators (lpe2B, lpe2C, lpe2D, lpe4A, lpe4B, and lpe4C).
In addition to these four clusters found on the chromosomal DNA, the lpe5 gene cluster, with a 22 kb size, is present on the plasmid pSN35N-3. As shown in Table 3 and Fig. 4A, the lpe5 cluster is composed of 16 ORFs (lpe5A–P) and contains 11 genes deduced to encode transposase around the cluster. Judging from the annotation information, the ORFs contained in the lpe5 cluster may be necessary for EPS biosynthesis, as well as the lpe2 and lpe4 clusters. However, we could not find a gene encoding phosphotransferase necessary for the phosphorylation of EPS in the cluster.
| Cluster and gene | Size (aa) | Location | Predicted function | Best BLAST match | Source organism | Accession No. | Identity (%) |
|---|---|---|---|---|---|---|---|
| lpe1 cluster | |||||||
| lpe1A | 323 | 593342–594313 C | Glycosyltransferase | Hypothetical protein | Lb. plantarum | WP_053338792 | 100 |
| lpe1B | 390 | 592170–593342 C | CDP-glycerol–glycerophosphate glycerophosphotransferase | Hypothetical protein | Lb. plantarum | WP_027821174 | 100 |
| lpe1C | 392 | 590311–591489 C | Glycosyltransferase | Glycosyltransferase family 1 protein | Lb. plantarum | WP_044430140 | 100 |
| lpe1D | 440 | 588992–590314 C | Glycosyltransferase | Hypothetical protein | Lb. plantarum | WP_053338793 | 100 |
| lpe2 cluster | |||||||
| lpe2A | 507 | 580323–581846 | Flippase Wzx | Transporter | Lb. plantarum | OBS43084 | 99.8 |
| lpe2B | 255 | 578507–579274 C | Chain-length determinant Wzz | Polysaccharide biosynthesis protein | Lb. plantarum | KWT43483 | 100 |
| lpe2C | 242 | 577767–578495 C | Tyrosine-protein kinase Wze | Exopolysaccharide biosynthesis protein | Lb. plantarum | WP_024971390 | 100 |
| lpe2D | 278 | 577007–577843 C | Protein-tyrosine phosphatase Wzb | Protein-tyrosine-phosphatase | Lb. plantarum | ARW35023 | 95.5 |
| lpe2E | 218 | 576301–576957 C | Priming glycosyltransferase | Sugar transferase | Lb. plantarum | WP_044430148 | 100 |
| lpe2F | 289 | 575219–576088 C | Glycosyltransferase | Hypothetical protein | Lb. plantarum | WP_044430150 | 100 |
| lpe2G | 418 | 573947–575203 C | Polymerase Wzy | Hypothetical protein | Lb. plantarum | WP_044430151 | 99.5 |
| lpe2H | 269 | 573134–573943 C | Glycosyltransferase | Hypothetical protein | Lb. plantarum | WP_053338795 | 99.6 |
| lpe2I | 237 | 572424–573137 C | Glycosyltransferase | Glycosyltransferase family 2 | Lb. plantarum | WP_044430157 | 100 |
| lpe2J | 473 | 569872–571293 C | Flippase Wzx | Hypothetical protein | Lb. plantarum | WP_080333751 | 100 |
| lpe3 cluster | |||||||
| lpe3A | 302 | 565206–566114 C | Glycosyltransferase | Glycosyl transferase family 2 | Lb. plantarum | WP_053338797 | 100 |
| lpe3B | 310 | 564240–565172 C | Glycosyltransferase | Glycosyl transferase family 2 | Lb. plantarum | WP_053338798 | 100 |
| lpe3C | 377 | 562594–563727 C | UDP-galactopyranose mutase | UDP-galactopyranose mutase | Lb. plantarum | WP_003644178 | 100 |
| lpe3D | 362 | 561445–562533 C | Tyrosine-protein kinase transmembrane module Wzd | Hypothetical protein | Lb. plantarum | WP_053338799 | 100 |
| lpe3E | 207 | 560816–561439 C | Tyrosine-protein kinase Wze | Hypothetical protein HMPREF0531_11724 | Lb. plantarum | EFK29287 | 100 |
| lpe3F | 406 | 559609–560829 C | Polymerase Wzy | Hypothetical protein | Lb. plantarum | WP_003644181 | 100 |
| lpe3G | 369 | 558503–559612 C | Unknown | Hypothetical protein | Lb. plantarum | WP_003644182 | 100 |
| lpe3H | 359 | 557440–558519 C | O-Acetyltransferase | Acetyltransferase | Lb. plantarum | WP_003644183 | 100 |
| lpe3I | 258 | 556528–557304 C | Glycosyltransferase | Exopolysaccharide biosynthesis protein | Lb. plantarum | WP_021356757 | 100 |
| lpe3J | 472 | 554920–556338 | Flippase Wzx | Flipplase | Lb. plantarum | WP_053338801 | 100 |
| lpe3K | 225 | 553806–554483 | Priming glycosyltransferase | Sugar transferase | Lb. plantarum | WP_075060689 | 99.6 |
| lpe4 cluster | |||||||
| lpe4A | 252 | 2975111–2975869 | Tyrosine-protein kinase transmembrane module Wzd | Polysaccharide biosynthesis protein | Lb. plantarum | WP_027821336 | 100 |
| lpe4B | 235 | 2975887–2976594 | Tyrosine-protein kinase Wze | Exopolysaccharide biosynthesis protein | Lb. plantarum | WP_003640787 | 100 |
| lpe4C | 273 | 2976533–2977354 | Protein-tyrosine phosphatase Wzb | Protein-tyrosine phosphatase | Lb. plantarum | CDN27632 | 100 |
| lpe4D | 313 | 2977370–2978311 | UDP-glucose 4-epimerase | Epimerase | Lb. plantarum | WP_053338960 | 100 |
| lpe4E | 221 | 2978298–2978963 | Priming glycosyltransferase | Capsular polysaccharide biosynthesis protein | Lb. plantarum | AOG30978 | 100 |
| lpe4F | 363 | 2978963–2980054 | Glycosyltransferase | Glycosyl transferase family 1 | Lb. plantarum | WP_053338958 | 99.7 |
| lpe4G | 342 | 2980070–2981098 | Glycosyltransferase | Glycosyl transferase family 1 | Lb. plantarum | WP_027822102 | 100 |
| lpe4H | 424 | 2981095–2982369 | Polymerase Wzy | Hypothetical protein | Lb. plantarum | WP_027822103 | 100 |
| lpe4I | 322 | 2982354–2983322 | Glycosyltransferase | Glycosyl transferase family 2 | Lb. plantarum | WP_027822104 | 100 |
| lpe4J | 324 | 2983879–2984853 | Flippase Wzx | Flipplase | Lb. plantarum | WP_027822105 | 100 |
| lpe5 cluster (on the plasmid pSN35N-3) | |||||||
| lpe5A | 374 | 979–2103 C | Glycosyltransferase | Glycosyltransferase family 2 protein | Bacillus coagulans | WP_051357575 | 42.9 |
| lpe5B | 485 | 7363–8820 C | Flippase Wzx | Flipplase | Lb. plantarum | WP_063487733 | 99.0 |
| lpe5C | 169 | 9977–10486 C | Glycosyltransferase | Glycosyl transferase | Bacillus cereus | WP_033687572 | 41.9 |
| lpe5D | 263 | 10483–11274 C | Glycosyltransferase | Hypothetical protein | Lb. plantarum | WP_080283862 | 46.1 |
| lpe5E | 302 | 11312–12220 C | Polymerase Wzy | EpsG family protein | Lb. vaginalis | WP_003717951 | 30.5 |
| lpe5F | 338 | 12446–13462 C | Glycosyltransferase | Glycosyltransferase family 2 protein | Clostridium clariflavum | WP_014255924 | 43.3 |
| lpe5G | 256 | 13452–14222 C | CDP-alcohol phosphatidyltransferas | Hypothetical protein | Lb. sakei | WP_082267650 | 57.2 |
| lpe5H | 149 | 14209–14658 C | Glycerol-3-phosphate cytidylyltransferase | Glycerol-3-phosphate cytidylyltransferase | Leuconostoc carnosum | WP_014974067 | 75.7 |
| lpe5I | 382 | 14661–15809 C | Glycosyltransferase | Hypothetical protein | Leuconostoc mesenteroides | WP_071952261 | 44.7 |
| lpe5J | 231 | 15836–16531 C | Priming glycosyltransferase | Sugar transferase | Lactobacillus coryniformis | WP_010014297 | 88.7 |
| lpe5K | 312 | 16578–17516 C | UDP-glucose 4-epimerase | UDP-glucose 4-epimerase | Lb. plantarum | WP_020923878 | 86.2 |
| lpe5L | 96 | 17541–17831 C | Protein-tyrosine phosphatase Wzb (truncated) | Tyrosine protein phosphatase | Lactobacillus sp. | WP_010495923 | 96.2 |
| lpe5M | 262 | 19596–20384 C | Protein-tyrosine phosphatase Wzb | Polysaccharide biosynthesis protein, phosphotyrosine-protein phosphatase | Lb. plantarum | AGS26747 | 98.5 |
| lpe5N | 242 | 20356–21084 C | Tyrosine-protein kinase Wze | Exopolysaccharide biosynthesis protein | Lb. plantarum | WP_072539917 | 99.2 |
| lpe5O | 255 | 21096–21863 C | Tyrosine-protein kinase transmembrane module Wzd | Chain length regulator | Lb. paraplantarum | CDF77689 | 98.0 |
| lpe5P | 154 | 22111–22575 C | Glycosyltransferase (truncated) | Hypothetical protein | Lb. collinoides | WP_063285095 | 100 |
To confirm whether genes for the biosynthesis of SN35N-derived EPS are present on the plasmid, Lb. plantarum SN35N cells were incubated with novobiocin used as a plasmid-curing agent. PCR analysis was carried out for a mutant that cannot produce the EPS. The result suggests that a DNA fragment containing lpe5J, which encodes a putative priming glycosyltransferase, in the EPS non-producing mutant SN35N-Δp3 was not detected. In addition, the EPS productivity of the mutant was drastically decreased (the EPS productivity of wild-type strain: 48.2 mg/L) until 3.3 mg/L. There results suggest that the lpe5 gene cluster present on the pSN35N-3 plasmid is involved in the production of the EPS.
Table 4 shows the sugar compositions of some Lb. plantarum strain-derived EPSs. EPS produced by Lb. plantarum 70810 is composed of only galactose as a monosaccharide.38) However, in the case of Lb. plantarum KF5, the produced EPS is composed of mannose, glucose, and galactose at a ratio of 1 : 4.99 : 6.9.39) Wang et al. have reported that the sugar components of EPS produced by Lb. plantarum YW32 were mannose, fructose, galactose, and glucose at a ratio of 8.2 : 1 : 4.1 : 4.2.40) According to another research group, the EPS produced by Lb. plantarum BC-25 was composed of glucose, galactose, and mannose at a molar ratio of 6.0 : 1.79 : 92.21.41) On the other hand, Lb. plantarum YW11, which has been isolated from Tibetan Kefir, produces EPS composed of glucose and galactose at a molar ratio of 2.7 : 1, with the possible presence of N-acetylated sugar residues in the polysaccharide.42) However, until now, the EPS-biosynthesizing gene cluster in both strains has not been analyzed.
| Strain | Composition sugars and ratio | Reference | ||||||
|---|---|---|---|---|---|---|---|---|
| Glucose | Galactose | Mannose | Fructose | Rhamnose | Glucosamine | Galactosamine | ||
| SN35N | 14.3 | 5.7 | 1.0 | — | — | — | — | This study |
| 70810 | — | 1.0 | — | — | — | — | — | 38 |
| KF5 | 4.99 | 6.9 | 1.0 | — | — | — | — | 39 |
| YW32 | 4.2 | 4.1 | 8.2 | 1.0 | — | — | — | 40 |
| BC-25 | 6.0 | 1.79 | 92.21 | — | — | — | — | 41 |
| YW11 | 2.7 | 1.0 | — | — | — | — | — | 42 |
| WCFS1 | 28 | 17 | — | — | 5.0 | 3.0 | — | 43 |
| SF2A35B | 3.0 | 66.6 | — | — | — | — | 313 | 43 |
| Lp90 | 3.9 | 22.2 | — | — | 25 | 24.5 | 24.4 | 43 |
On the other hand, the EPS-biosynthesizing gene clusters for the WCFS1, SF2A35B, and Lp90 strains, which belong to Lb. plantarum, have been analyzed together with the EPS components.43) The EPS from the WCFS1 strain contains glucose, galactose, glucosamine, and rhamnose at a ratio of 28 : 17 : 3 : 5. The SF2A35B strain produces an EPS composed of galactose and galactosamine, but little glucose (66.6 : 313 : 3, respectively). The monosaccharide component ratio of EPS produced by Lb. plantarum Lp90 is composed of glucosamine, galactose, galactosamine, and rhamnose at almost the same, but with a small ratio of glucose (24.5 : 22.2 : 24.4 : 25.0 : 3.9). Our present study reveals that SN35N-derived EPS consists of glucose, galactose, and mannose moieties at a ratio of 15.0 : 5.7 : 1.0. Thus, the molar ratios of the composed monosaccharides are significantly different, even among the same species.
It has been reported that EPSs produced by the Lb. amylovorus PY45 and P. pentosaceus LY45 inhibit the catalytic activity of hyaluronidase.22) The phenomenon correlates with the inhibition of histamine release in inflammatory reactions accompanied by the immunoglobulin E-mediated mast cell degranulation.44,45) The present study shows that the IC50 value for the SN35N-derived acidic EPS to hyaluronidase has almost the same as those of sodium cromoglicate and dipotassium glycyrrhizinate used as an anti-inflammatory agent (hyaluronidase inhibitors).45,46) Interestingly, although our previous report has showed that the neutral EPS produced by P. pentosaceus LY45 inhibits hyaluronidase activity more effectively than did the acidic EPS, including fucoidan.22) The IC50 value of the SN35N-derived acidic EPS is significantly lower than those of the acidic ones. In addition, the mixture of neutral and acidic EPSs may inhibit hyaluronidase activity synergistically. The synergistic inhibitory activity may be due to its structure or composition rather than its acidic properties.
A whole-genome analysis of the SN35N strain demonstrates that there are four possible EPS-biosynthetic gene clusters, designated lpe1 to lpe4, on the chromosomal DNA (Table 3, Figs. 3, and 4). As compared with the Lb. plantarum WCFS1, although the lpe1 cluster is not conserved, three other clusters are significantly conserved, especially in the lpe4 cluster, against the cognate clusters designated cps1–4 in WCFS1 (Fig. 3). Based on a homology search, the lpe2 and lpe4 clusters may be expected to function as intact gene clusters.

The gene organizations and comparisons of the gene clusters among four Lb. plantarum strains are indicated. The gray connecting areas indicate highly conserved regions among the indicated strains.
Although four EPS biosynthesis gene clusters are present on the Lb. plantarum SN35N chromosomal DNA, as shown in the result of the plasmid-curing experiment, it is suggested that the lpe5 cluster found on the plasmid pSN35N-3 mainly participates in the production of SN35N-derived acidic EPS. Furthermore, it has been shown that some Lactobacillus strains, such as Lb. paracasei DG,47) Lb. casei YIT9018,48) and Lb. rhamnosus RW-9595M,49) have four genes, designated rmlA–D, which are necessary for synthesis of rhamnose as a component of the EPS in their EPS-biosynthetic gene clusters. In fact, these three strains produce EPS that contains rhamnose. However, rmlA–D genes are between the lpe1 and lpe2 clusters found on SN35N chromosomal DNA, whereas rhamnose was not detected as a component of the EPS from the SN35N strain. The results indicate that at least rmlA–D genes do not contribute to EPS production by Lb. plantarum SN35N. Our present results suggest that acidic EPS produced by the SN35N strain is phosphorylated EPS. However, a gene for the phosphorylation of EPS is not found near or in the EPS-biosynthesizing gene cluster.
Remus et al. investigated the deletion mutants of each cps gene cluster on Lb. plantarum WCFS150) and showed that two of the four cps clusters contain all of the genes required for polysaccharide formation (cps2A–J and cps4A–J). They demonstrated that all cps clusters function in the WCFS1 strain, despite of the incompleteness of other cps1 and cps3 clusters. This result shows the possibility that the existence of WCFS1 chromosome-encoded lpe1–4 clusters is somehow connected to the structure and component sugars of the WCFS1-produced EPS.
In the present study, the EPS-biosynthesizing gene cluster found in the SN35N strain was compared with those in the WCFS1, SFA35B, and Lp90 strains (Fig. 3). The gene disruption experiment has suggested that ORFs for the biosynthesis of EPS encoded in the WCSF1 strain are present in the deduced EPS-synthesizing genes, designated cps1, cps2, cps3, and cps4.50) In the case of the SF2A35B and Lp90 strains, it has been shown that the production of EPS was scarcely detected when the cps2-like gene was disrupted.43) As shown in the present study, although the lpe2–lpe4 clusters homologous to the cps2–cps4 clusters are found on the SN35N genome, no EPS productivity was detected when the plasmid carrying the lps5 gene cluster was cured from the SN35N cell by treatment with novobiocin. This result suggests that the EPS-biosynthesizing gene cluster is strain specific, even in the same Lactobacillus species. That is, even if several EPS-biosynthesizing gene clusters are found in Lb. plantarum strains, the EPS gene cluster for EPS production is strain specific.
Lb. buchneri CD034 and Lactococcus lactis NIZO B20, harboring plasmids designated pCD034-351) and pNZ4000,52) respectively, have been reported to carry the EPS-biosynthesizing gene cluster, such as a Lb. plantarum SN35N plasmid pSN35N-3. The NCBI database suggests that six strains of Lb. plantarum may harbor the plasmid carrying the putative EPS-biosynthesizing gene cluster (Fig. 4B). However, studies confirming the phenomenon have not existed until now.
Interestingly, several transposase-encoding genes are found on the lpe5 gene cluster located on the Lb. plantarum SN35N plasmid, pSN35N-3. At one time, the EPS-synthesizing gene cluster present chromosomally may have been responsible for the transposable integration into pSN35N-3. Table 3 shows that some lpe5 cluster-encoded genes are homologous with that found in the genera Leuconostoc and Bacillus, whereas the lpe1, lpe2, ple3, and lpe4 ones are homologous only with the species Lb. plantarum. This result indicates that the lpe5 gene cluster is actively involved in the production of SN35N-specific EPS.

(A) Physical map and gene organization of plasmid pSN35N-3 (accession no. AP018408). Gene organization in putative EPS biosynthesis cluster lpe5 is shown as a gray-shaded area. (B) The plasmid-encoded putative EPS biosynthesis gene clusters found on the Lb. plantarum strains are indicated with strain names as follows: strains 16 (accession no. NC_021514), C410L1 (NZ_CP017954), HFC8 (NZ_CP012650), LZ227 (NZ_CP015857), TMW1.1623 (NZ_CP017379), and ZJ316 (NC_020229).
In recent years, the health benefits of natural polysaccharides, such as fucoidan and β-glucan, have drawn considerable attention.53,54) Furthermore, as with other LAB species, there are some reports of a few strains of Lb. plantarum producing functional EPSs, such as radical scavengers, dendritic cell maturation induction, and anti-bacterial activities.55–57)
The investigation of physiological functions of the SN35N-derived EPS, such as anti-allergy, anti-helicobacter pylori, anti-ulcer, and anti-viral activities, is in progress.
We thank the Research Center for Molecular Medicine, the Faculty of Medicine, and the Analysis Center of Life Science, Hiroshima University, for the use of their facilities. We are also grateful to Mr. Naoto Kaji, Kohjin Life Sciences Co., Ltd., Oita, Japan, for his collaboration on the early stages of this work.
The authors declare no conflict of interest.
The GenBank/EMBL/DDBJ accession numbers for the sequences reported in this paper are AP018405, AP018406, AP018407, AP018408, and AP018409.