2019 年 42 巻 5 号 p. 827-832
2019 年 42 巻 5 号 p. 827-832
Human parainfluenza virus type 1 (hPIV1) has two spike glycoproteins, the hemagglutinin-neuraminidase (HN) glycoprotein as a receptor-binding protein and the fusion (F) glycoprotein as a membrane-fusion protein. The F glycoprotein mediates both membrane fusion between the virus and cell and membrane fusion between cells, called syncytium formation. Wild-type C35 strain (WT) of hPIV1 shows little syncytium formation of infected cells during virus growth. In the present study, we isolated a variant virus (Vr) from the WT that showed enhanced syncytium formation of infected cells by using our previously established hPIV1 plaque formation assay. Vr formed a larger focus and showed increased virus growth compared with WT. Sequence analysis of the spike glycoprotein genes showed that the Vr had a single amino acid substitution of Ile to Val at position 131 in the fusion peptide region of the F glycoprotein without any substitutions of the HN glycoprotein. The Vr F glycoprotein showed enhanced syncytium formation in F and HN glycoprotein-expressing cells. Additionally, expression of the Vr F glycoprotein increased the focus area of the WT-infected cells. The single amino acid substitution at position 131 in the F glycoprotein of hPIV1 gives hPIV1 abilities to enhance syncytium formation and increase cell-to-cell spread. The present study supports the possibility that hPIV1 acquires increased virus growth in vitro from promotion of direct cell-to-cell transmission by syncytium formation.
Human parainfluenza virus type 1 (hPIV1) is a respiratory pathogen that causes bronchiolitis, pneumonia, laryngotracheobronchitis and croup in infants or young children.1–4) hPIV1 infections may also cause more severe respiratory diseases, especially in the elderly and immunocompromised patients. However, there is no clinical therapy or vaccine against hPIV1.5) hPIV1 has two spike glycoproteins, hemagglutinin-neuraminidase (HN) glycoprotein as a receptor-binding protein and fusion (F) glycoprotein as a membrane-fusion protein, on the viral surface.6) hPIV1 binds to terminal sialic acids of glycoconjugates on the host cell surface through the HN glycoprotein.6,7) The F glycoprotein is a common spike protein in paramyxovirus and mediates both membrane fusion between the virus and cell (virus–cell fusion) and membrane fusion between cells (cell–cell fusion).8)
Nascent HN and F glycoproteins are abundantly expressed on the cell surface membrane of paramyxovirus-infected cells. The F glycoprotein on the cell surface membrane induces cell–cell membrane fusion between virus-infected cells and their neighboring non-infected cells, resulting in multinucleated giant cell formation, called syncytium formation.1) Paramyxovirus simian virus 5 spreads infection with cell-to-cell direct transmission by syncytium formation.9) Syncytium formation has been reported to be associated with paramyxovirus growth.8,10) Remarkable syncytium formation can be seen in cells infected with some paramyxoviruses such as Sendai virus (SeV), respiratory syncytial virus, and human parainfluenza virus type 3 (hPIV3).11,12) hPIV1 shows little syncytium formation during virus growth in vitro.8,13)
We previously established a hPIV1 plaque formation assay.14) In the present study, we isolated an hPIV1 variant (Vr) that had acquired enhanced syncytium formation ability compared with wild-type C35 strain (WT) of hPIV1 by our plaque formation assay. The Vr showed increased progeny virus release and formed larger plaques than did WT. The Vr had a single amino acid substitution of Ile to Val at position 131 in the fusion peptide region of the F glycoprotein without any substitutions in the HN glycoprotein. The single amino acid substitution at position 131 of the F glycoprotein enhanced syncytium formation and increased the focus sizes. This study showed that the single amino acid substitution at position 131 in the fusion peptide region of the F glycoprotein of hPIV1 gives hPIV1 abilities to enhance syncytium formation and increase cell-to-cell spread in vitro.
Lewis lung carcinoma-monkey kidney (LLC-MK2) cells were maintained in Eagle’s minimum essential medium (MEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS). Human epidermoid cancer (Hep-2) cells were maintained in Dulbecco’s modified Eagle’s MEM supplemented with 10% FBS. The WT (C35 strain) of hPIV1 was kindly provided by Allen Portner of St. Jude Children’s Research Hospital. The WT was propagated in LLC-MK2 cells. Rabbit anti-hPIV1 antibody was prepared by immunization with the WT as described previously and was purified on a Protein G HiTap™ Protein G HP (CE Healthcare Life Sciences, Piscataway, NJ, U.S.A.).15)
Isolation of a Variant Virus Showing Obvious Syncytium FormationThe plaque assay of the WT was carried out as in our previous study using confluent monolayers of LLC-MK2 cells.14) A plaque in which obvious syncytium formation was observed under a microscope was selected, and then the Vr was obtained from the plaque and was inoculated into newly confluent monolayers of LLC-MK2 cells in a 6-well plate. The cells were incubated in hybridoma-serum-free medium complete dry powdered medium (SFM, Thermo Fisher Scientific Inc., Waltham, MA, U.S.A.) containing 3 µg/mL of acetylated trypsin at 37°C for 3 d. The culture medium was harvested and was added to newly confluent monolayers of LLC-MK2 cells in a 6-well plate. The cells were incubated in SFM containing 3 µg/mL of acetylated trypsin at 37°C for 3 d again. The plaque assay was again performed with the harvested culture medium. Finally, the Vr showing obvious syncytium formation was isolated from the plaque formed by the WT.
Construction of an Expression Plasmid Vector Containing F or HN GeneF genes and HN genes were cloned into the pCAG GS/MCS expression vector described previously.8) Briefly, each viral RNA was extracted from virus particles with TRIzol reagent (Invitrogen Corp., Carlsbad, CA, U.S.A.) according to the manufacturer’s instructions. Full-length cDNAs of the F gene and HN gene from hPIV1 were amplified by using a TaKaRa RNA PCR™ kit Ver. 3.0 (TaKaRa Bio Inc., Shiga, Japan) with the primers described in Supplementary Table 1. The F genes and HN genes were inserted into the multiple cloning region between the EcoRI site and XhoI site of the pCAG GS/MCS expression plasmid vector. Sequence analysis of the F gene and the HN gene was performed by using ABI Prizm 310 Genetic Analyzer (Life Technologies, Calsbad, CA, U.S.A.) with the primers shown in Supplementary Table 2, and the nucleotide substitution of the Vr was identified.
Focus-Forming Assay and Virus TitrationConfluent monolayers of LLC-MK2 cells in a 12-well plate were washed with phosphate buffered saline (PBS) and were inoculated with serial dilutions of a virus suspension at room temperature for 30 min. The wells were washed with PBS and the cells were overlaid with 2 mL of SFM containing 3 µg/mL acetylated trypsin and 1.2% microcrystalline cellulose Avicel (FMC Corporation, Philadelphia, PA, U.S.A.) at 37°C for 2 d. The cells were fixed with 800 µL of cold methanol for 5 min and reacted with rabbit anti-hPIV1 antibody at room temperature for 30 min and then with horseradish peroxidase-conjugated Protein A (Sigma-Aldrich, St. Louis, MO, U.S.A.) at room temperature for 30 min. Focus-forming units (FFU) as infectious virus titers were calculated from the immunostained foci developed as described previously.16)
A focus-forming assay of WT was also performed in F glycoprotein-expressing cells. Seventy percent confluent monolayers of LLC-MK2 cells in a 12-well plate were transfected with 1 µg of pCAG GS vectors containing the F gene from WT (F-WT gene) or F gene from Vr (F-Vr gene) using the transfection reagent TransIT-LT1 (Mirus, Madison, WI, U.S.A.) according to the manufacturer’s instructions. Cells were also mock-treated without plasmid vectors as a control. After incubation at 37°C for 6 h, the transfected cells were washed with PBS and then the cells were incubated with 1 mL of SFM. After incubation for 22 h, the transfected cells were washed with PBS and the focus-forming assay was performed with WT at a multiplicity of infection (MOI) of 0.0001 for 3 d incubation. The area of the stained focus was measured using Image J release 1.40 g (National Institutes of Health, U.S.A., http://rsb.info.nih.gov/ij/) and represented as the relative focus area to the median of the control focus areas.
Estimation of Multi-Step Growth of the VirusConfluent monolayers of LLC-MK2 cells on a 24-well plate were infected with viruses at a MOI of 0.001 for 30 min at room temperature. After washing the viruses with 250 µL of PBS, the cells were incubated with 500 µL of SFM containing 3 µg/mL of acetylated trypsin. Supernatants of the cells were recovered at 12, 24 and 36 h. Virus titers in the supernatants were measured by a focus-forming assay with triplicated experiments.
Syncytium Formation AssayThe hPIV1 syncytium formation assay (cell-cell fusion assay) was carried out as in our previous study.8) Briefly, for quantification of syncytium formation of hPIV1-infected cells, confluent monolayers of LLC-MK2 cells in a 24-well plate were infected with each virus in SFM at an MOI of 1 at room temperature for 30 min. The plates were incubated at 37°C for 48 h in culture of SFM containing 3 µg/mL of acetylated trypsin until visualization of syncytium formation. The infected cells were fixed with 500 µL of cold methanol for 5 min and then treated with 500 µL of Carrazzi’s haematoxylin solution in PBS (1 : 1, v/v) (Wako Pure Chemical Industries, Ltd., Osaka, Japan) in order to visualize the nuclei. Fusion efficiency (%) of the infected cells was calculated as percentage of the number of nuclei in the syncytia relative to the sum total of nuclei in all cells under 9 optical observations at a magnification of 100.
For quantification of syncytium formation of LLC-MK2 cells expressing both F and HN genes, 70% confluent monolayers of LLC-MK2 cells in a 24-well plate were co-transfected with 1 µg of both pCAG GS vectors containing F and HN genes using the transfection reagent TransIT-LT1 according to the manufacturer’s instructions. After incubation at 37°C for 6 h, the co-transfected cells were washed with PBS and then the cells were incubated with 500 µL of SFM containing 3 µg/mL of acetylated trypsin. After incubation for 46 h, the co-transfected cells were washed with PBS. Fusion efficiency (%) of the co-transfected cells was calculated as percentage of the number of nuclei in the syncytia relative to the sum total of nuclei in all cells under 5 optical observations at a magnification of 100.
For quantification of syncytium formation of Hep-2 cells expressing both F and HN genes, 70% confluent monolayers of Hep-2 cells in a 24-well plate were co-transfected with 500 ng of both pCAG GS vectors containing F and HN genes using the transfection reagent according to the manufacturer’s instructions. After incubation at 37°C for 48 h, the co-transfected cells were treated with 3 µg/mL of acetylated trypsin at 37°C for 30 min. The cells were washed with PBS and incubated with 500 µL of SFM. After incubation for 10 h, the co-transfected cells were washed with PBS. Fusion efficiency (%) of the co-transfected Hep-2 cells was calculated as percentage of the number of nuclei in the syncytia relative to the sum total of nuclei in all cells under 5 optical observations at a magnification of 100.
We previously established a hPIV1 plaque formation assay.14) hPIV1 generally shows little syncytium formation in vitro.8,13) However, in our plaque formation assay of hPIV1, there were a few plaques in which obvious syncytium formation was observed (a few syncytium-forming plaques per 100 non-syncytial plaques). It seemed that hPIV1 fortuitously acquired mutations that induced enhanced syncytium formation, by continuously passing the virus in LLC-MK2 cells. We isolated a variant virus (Vr) showing enhanced syncytium formation by selecting a syncytium-forming plaque. To compare the syncytium formation activities of the Vr and the parent C35 strain of hPIV1 (WT), LLC-MK2 cells were infected with each virus at an MOI of 1 and incubated in the presence of acetylated trypsin until clear appearance of syncytia. Whereas syncytium formation was hardly observed in the WT-infected cells, obvious syncytium formation was observed in the Vr-infected cells (Fig. 1A). The Vr showed approximately 4-times higher syncytium formation activity than that of the WT (Fig. 1B).
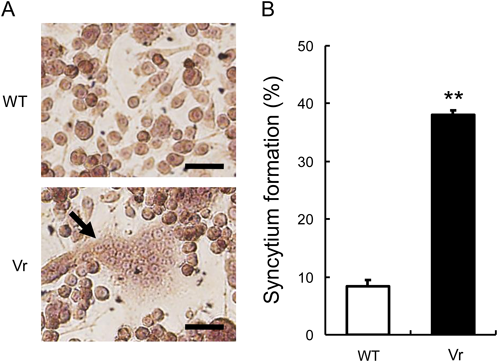
LLC-MK2 cells were infected with the WT or the Vr at an MOI of 1 and incubated for 48 h. Nuclei were stained with hematoxylin. (A) Observation of syncytium formation of WT-infected cells and Vr-infected cells. The arrows indicate remarkable syncytium formation. Scale bars indicate 50 µm. (B) Syncytium formation (%) of the infected cells in (A) was calculated as described in MATERIALS AND METHODS. Totally, 5939 nuclei (WT) and 6184 nuclei (Vr) were counted. ** p < 0.01 versus the result for the WT, unpaired Student’s t test. (Color figure can be accessed in the online version.)
It has been reported that amino acid substitutions of the viral spike HN and F glycoproteins enhance fusion activity of SeV, hPIV1, and hPIV3.8,12,17) We therefore compared amino acid sequences of the HN glycoprotein and the F glycoprotein between the Vr and the WT. The results revealed that the Vr carried a single amino acid substitution of Ile to Val at position 131 in the F glycoprotein without any amino acid substitutions in the HN glycoprotein. According to the amino acid residue alignment with the F glycoprotein from parainfluenza virus type 5 (PIV5) whose three-dimensional structure has been determined,18) the amino acid position 131 of F glycoprotein of hPIV1 was located in the fusion peptide region which is directly inserted into the target membrane for the membrane fusion process. To determine whether the single amino acid substitution in the F glycoprotein of the Vr enhances syncytium formation of hPIV1, the F gene of the WT (F-WT gene) or the Vr (F-Vr gene) was co-transfected together with the HN gene of the WT (HN-WT gene) into LLC-MK2 cells. The transfected cells were incubated in the presence of acetylated trypsin until clear appearance of syncytia. The cells with the HN-WT glycoprotein and the F-WT glycoprotein showed less syncytium formation. On the other hand, the cells with the HN-WT glycoprotein and the F-Vr glycoprotein showed approximately 2-times higher syncytium formation activity than that of the cells with the HN-WT and F-WT glycoproteins (Fig. 2A). Transfection with each F gene alone without the HN gene induced no syncytium formation, indicating that receptor binding activity of the HN glycoprotein is needed for the membrane fusion process of F-WT and F-Vr glycoproteins. For Hep-2 cells, which show obvious syncytium formation in hPIV3 infection, co-transfection with the F-Vr gene together with the HN-WT gene also induced approximately 8-times higher syncytium formation activity than that with HN-WT and F-WT genes (Fig. 2B). These results demonstrated that the amino acid substitution of Ile to Val at position 131 in the hPIV1 F glycoprotein gave enhanced syncytium formation ability in different cell lines, LLC-MK2 and Hep-2 cells. It seemed that expression of HN-WT and F-WT glycoproteins showed less syncytium formation in Hep-2 cells than in LLC-MK2 cells. In our previous study, we used LLC-MK2 cells and Hep-2 cells for establishment of a WT hPIV1 plaque assay.14) Obvious plaque formation is dependent largely on cell death and/or syncytium formation of infected cells. We succeeded in achieving WT plaque formation in LLC-MK2 cells but not in Hep-2 cells. For WT infection, Hep-2 cells showed lower rates of cell death and syncytium formation than did LLC-MK2 cells. It is thought that Hep-2 cells have less fusion efficiency in the case of WT hPIV1 infection.
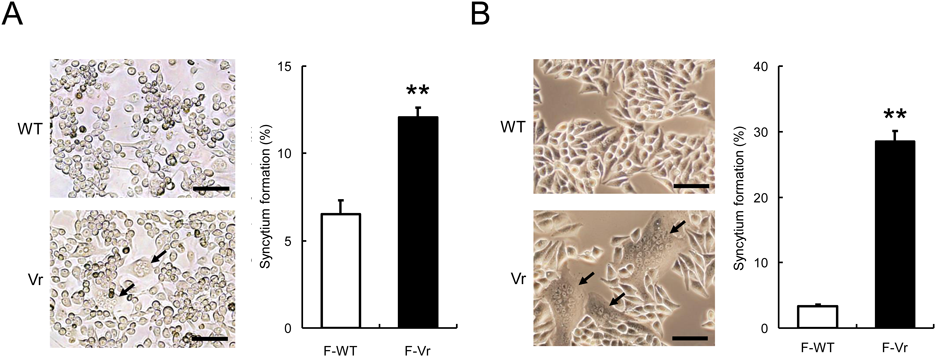
(A) Fusion efficiency of LLC-MK2 cells co-transfected with the F genes together with the WT HN gene. The cells were co-transfected with the respective F gene from the WT or the Vr together with the WT HN gene. After incubation for 46 h, syncytium formation (%) was calculated as described in MATERIALS AND METHODS. Totally, 4616 nuclei (F-WT) and 5111 nuclei (F-Vr) were counted. (B) Fusion efficiency of Hep-2 cells co-transfected with the F genes together with the WT HN gene. The cells were co-transfected with the respective F gene from the WT or the Vr together with the WT HN gene. After incubation for 58 h, syncytium formation (%) was calculated as described in the MATERIALS AND METHODS section. Totally 4135 nuclei (F-WT) or 4829 nuclei (F-Vr) were counted. Scale bars indicate 40 µm. The arrows indicate remarkable syncytium formation. ** p < 0.01 versus the result for the WT, unpaired Student’s t test. (Color figure can be accessed in the online version.)
We compared the amino acid sequences near the position 131 of F glycoprotein among 9 kinds of paramyxoviruses, including the hPIV1 WT and the Vr. The amino acid residue at the position 131 is Ile in hPIV1 WT, human parainfluenza virus type 2, hPIV3, SeV, and Newcastle disease virus, or Val in hPIV1 Vr, human parainfluenza virus type 4, PIV5, and mumps virus. Among these viruses, the fusion peptide region between 113 and 135 is highly conserved19) (Fig. 3). For paramyxoviruses other than hPIV1, the I131V substitution is also suggested to increase fusion activity of each F glycoprotein, which enhances syncytium formation.

Amino acid sequences of F glycoprotein of hPIV1 WT (GenBank accession number AAA46800), hPIV1 Vr, human parainfluenza virus type 2 (hPIV2, GenBank accession number AAA46842), hPIV3 (GenBank accession number AAA46841), human parainfluenza virus type 4 (hPIV4, GenBank accession number BAA08626), PIV5 (GenBank accession number AAA47881), SeV (GenBank accession number AAB06281), Newcastle disease virus (NDV, GenBank accession number ACO25492) and mumps virus (MuV, GenBank accession number ABF71366) were compared by CLUSTALW (http://align.genome.jp/). The hatching sequences indicate fusion peptide region. Number shows amino acid numbering based on hPIV1.
It has been reported that enhanced syncytium formation activity promotes multi-step growth of hPIV18) and cell-to-cell spread of Simian virus 5.9) To evaluate virus growth of the Vr, we estimated the focus size as cell-to-cell spread of infection and quantified the supernatant virus titer of infected cells as multi-step growth. Focus sizes of the Vr were significantly larger than those of the WT (Figs. 4A, B). The Vr also significantly increased progeny virus titer in the culture supernatant of infected LLC-MK2 cells during multi-step growth compared to that in the case of the WT (Fig. 4C). These results showed that an enhanced syncytium-forming hPIV1 variant, Vr, increased cell-to-cell spread and multi-step growth in vitro.
(A) Focus images of the WT and the Vr. (B) Comparison of focus areas between the WT (n = 63) and Vr (n = 41). Focus areas were calculated and represented as the relative focus area to the median of the WT focus areas. Bars indicate each median. ** p < 0.01 versus the result for WT, Mann–Whitney U test. (C) Comparison of multi-step growth between the WT (open square) and Vr (closed circle). * p < 0.05; ** p < 0.01 versus the result for WT, unpaired Student’s t test. (Color figure can be accessed in the online version.)
To examine whether the single amino acid substitution of Ile to Val at position 131 in the F-Vr glycoprotein affects cell-to-cell spread of infection, we measured focus sizes when F-Vr glycoprotein-expressing cells were infected with the WT. The promotive effect of syncytium formation on direct cell-to-cell transmission of hPIV1 should result in larger focus sizes. LLC-MK2 cells were transfected with the F-WT gene or the F-Vr gene and then infected with the WT. After culture for 3 d, focus sizes were compared. Both F-WT glycoprotein expression and F-Vr glycoprotein expression increased focus areas of the WT compared to that with no transfection of F gene. F-Vr glycoprotein expression resulted in a statistically significant enlargement of focus areas of the WT compared to that with F-WT glycoprotein expression (Fig. 5). The results showed that a single amino acid substitution of Ile to Val at position 131 in the hPIV1 F glycoprotein increased cell-to-cell spread of hPIV1 in vitro.
LLC-MK2 cells were transfected with the F-WT gene or the F-Vr gene. The cells were also mock-treated without a plasmid. Ctrl means control experiment without a plasmid. After incubation for 28 h, focus-forming assay of the WT virus was performed in F glycoprotein-expressing cells. Focus areas were calculated and represented as the relative focus areas to the median of Ctrl focus areas (F-WT, n = 43; F-Vr, n = 35; Ctrl, n = 76). Bars indicate each median. ** p < 0.01 versus the result for F-WT; # p < 0.05; ## p < 0.01 versus the result for Ctrl, Steel–Dwass’ test.
In the present study, we isolated a Vr showing enhanced syncytium formation ability and increased virus growth in vitro by using our established hPIV1 plaque formation assay. The Vr is thought to have appeared by natural occurrence during several passages of the WT. We identified the single amino acid substitution of Ile to Val at position 131 of the fusion peptide region in the F glycoprotein carried by the Vr. The single amino acid substitution of the F glycoprotein gave the hPIV1 abilities to enhance syncytium formation and to increase cell-to-cell spread of virus infection in vitro. The present study supports the possibility that hPIV1 acquires increased virus growth from promotion of direct cell-to-cell transmission by syncytium formation.
Keijo Fukushima was a recipient of a scholarship by Honjo International Scholarship Foundation. This work was supported by JSPS KAKENHI, Series of single-year Grants, 2510690, in part, and by THE ICHIRO KANEHARA FOUNDATION, a JSPS KAKENHI Grant (Scientific Research C, JP23590549; Challenging Exploratory Research, JP26670064, JP16K15151; Young Scientists A, JP15H05644; Young Scientists B, JP18K14905).
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.