2020 年 43 巻 5 号 p. 823-830
2020 年 43 巻 5 号 p. 823-830
Efficiency (speed and cost) and animal welfare are important factors in the development of new drugs. A novel method (the half-life method) was developed to predict the human plasma concentration–time profile of a monoclonal antibody (mAb) after intravenous (i.v.) administration using less data compared to the conventional approach; moreover, predicted results were comparable to conventional method. This new method use human geometric means of pharmacokinetics (PK) parameters and the non-human primates (NHP) half-life of each mAb. PK data on mAbs in humans and NHPs were collected from literature focusing on linear elimination, and the two-compartment model was used for analysis. The following features were revealed in humans: 1) the coefficient of variation in the distribution volume of the central compartment and at steady state of mAbs was small (22.6 and 23.8%, respectively) and 2) half-life at the elimination phase (t1/2β) was the main contributor to plasma clearance. Moreover, distribution volume showed no significant correlation between humans and NHPs, and human t1/2β showed a good correlation with allometrically scaled t1/2β of NHP. Based on the features revealed in this study, we propose a new method for predicting the human plasma concentration–time profile of mAbs after i.v. dosing. When tested, this half-life method showed reasonable human prediction compared with a conventional empirical approach. The half-life method only requires t1/2β to predict human PK, and is therefore able to improve animal welfare and potentially accelerate the drug development process.
Non-clinical pharmacokinetic studies are conducted to predict human pharmacokinetics (PK) for drug development.1) In addition to efficiency, such as speed and cost, animal welfare is another important aspect in recent circumstances.2) Unfortunately, blood must be drawn and sampled more than 10 times in non-human primates (NHP) studies for PK analysis.3,4) A method of predicting human PK that requires less data would be preferable, both in terms of efficiency and animal welfare.5)
Progress in technology related to recombinant DNA and hybridomas for monoclonal antibodies (mAbs) has increased the use of active peptides, proteins, and mAbs for therapeutic purposes.6,7) A significant feature of these biologics is that their interactions with a specific receptor or antigen mediate their distribution and elimination.8) Active peptides, such as erythropoietin (EPO) and granulocyte-colony stimulating factor (G-CSF), are cleared from circulating blood not only by receptor-mediated (non-linear) elimination but also by linear elimination, for which the linear clearance (CL) can be predicted by allometric scaling.9,10) In recent years, numerous therapeutic mAbs have come on the market, and new candidates are continuously being developed for cancer, autoimmune disorders, and other diseases.11,12) As well as binding to other biologics, mAbs can also bind to membrane antigen, in which case their elimination processes are both non-linear and linear.13) Antibodies can avoid cellular degradation by recycling the mechanisms of human neonatal Fc receptor (FcRn) and, therefore, have a long circulating half-life in healthy humans (7–21 d).14) The prediction of a human plasma concentration–time profile with non-linear elimination is difficult and complex, requiring the inclusion of numerous factors, e.g., the target antigen and mAb-target complex turn-over rate, the antigen synthesis rate, and mAb affinity to the target antigen.15) For these reasons, we focused on predicting linear elimination in this report.
In a report by Dirks and Meibohm that reviewed the human population PK parameters of therapeutic mAbs, the PK data exhibited a bi-exponential decay pattern of linear elimination after intravenous (i.v.) administration, and a number of mAbs were analyzed by the two-compartment model.16) Their report indicated that, because the distribution of mAbs is restricted by their large molecular weight and high hydrophilicity, the distribution volume at central (V1) and at peripheral (V2) were typically small in humans.
This similarity in the PK features suggests that some values for PK parameters of mAbs may be common in humans. If so, these parameters would not need to be extrapolated from NHP values. Therefore, in this study, the human PK parameters of mAbs in published data were collected and analyzed to determine whether parameters extrapolated from NHPs are needed to predict the human plasma concentration–time profile after i.v. dosing.
Predictions about human CL and plasma concentration–time profile are an important part of drug development, especially prior to first-in-human clinical studies.17) Predicting human CL by allometry has been reported using multiple species,18,19) including cynomolgus monkeys,20–23) transgenic mice expressing human FcRn,24,25) and minipigs.26) Of these species, the cynomolgus monkey is the most frequently selected for preclinical studies to predict human CL. In addition to human CL, human plasma concentration–time profiles have also been predicted, both empirically and mechanistically. As shown in Fig. 1, examples of empirical approaches are allometric scaling of two-compartment model parameters,13,27) and Dedrick plot28,29) while an example of the mechanistic approach is the physiologically based pharmacokinetics model (PBPK).30–32) However, an empirical approach based on human PK for predicting human plasma concentration–time profile of mAbs after i.v. dosing has not been reported. This report proposes the half-life method of prediction in humans, using human geometric means and the allometric scaled t1/2β of NHP.

The main search objectives were therapeutic mAbs approved by the FDA (Food and Drug Administration, U.S.A.) or PMDA (Pharmaceutical and Medical Devices Agency, Japan) before the end of 2015. Moreover, some agents under clinical development were also added to the dataset, which consisted of data on human PK after i.v. administration (24 mAbs),33–55) cynomolgus monkey PK after i.v. administration (15 mAbs),4,33,39,42,44,49,51,55–61) and rhesus monkey PK after i.v. administration (4 mAbs).3,52,62,63) The PK of mAbs with linear PK in both humans and NHPs were analyzed and the parameter values of the selected mAbs are listed in the table in Supplementary 1. When two-compartment model parameters could not be obtained from literature, two-compartment model analysis was conducted on the data captured for serum/plasma mean concentration versus time using digitizer software UnGraph Version 5.0 (HULINKS, Tokyo, Japan).
Pharmacokinetic Data AnalysisThe serum/plasma mean concentration–time profiles after i.v. administration were analyzed by the two-compartment open model using Phoenix WinNonlin version 6.0 (Certara Inc., Princeton NJ, U.S.A.). The serum/plasma concentration (C) declined in a bi-exponential fashion using first macro-constant (A), second macro-constant (B), first rate constant (α), and second rate constant (β).
 | (1) |
Central elimination rate (K10), central to peripheral distribution rate (K12), and peripheral to central distribution rate (K21) were obtained according to the following equations:
 | (2) |
 | (3) |
 | (4) |
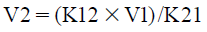 | (5) |
The distribution volume in central compartment (V1) was calculated as Dose/(A + B); the distribution volume at steady state (Vdss) was calculated as V1 + V2; t1/2β was calculated as Ln (2)/β.
The AUC was calculated by the following equations:
 | (6) |
 | (7) |
 | (8) |
The total body CL was estimated as Dose/AUCinf, and the distribution volume at the elimination phase (Vdβ) was calculated as CL/β.
Prediction of Human Elimination Rate Constants in the Elimination Phase Using Allometric ScalingThe human β of mAbs was predicted based on NHP data using the simple allometric equation:
 | (9) |
βhuman and βNHP are elimination rates at the distribution phase in humans and NHPs, respectively. BWhuman is the human body weight and BWNHP is the cynomolgus monkey or rhesus monkey body weight. If no specific body weight information was found in literature, a weight of 70 kg for humans, 3.75 kg for cynomolgus monkeys, or 6.0 kg for rhesus monkeys was used.20) X is the scaling exponent for rate constant (−0.15).21)
Predicting Human Plasma Concentration–Time Profiles by the Half-life MethodThe human plasma concentration–time profiles after i.v. dosing were predicted from NHP t1/2β. For the half-life method, the human geometric mean values for V1human, Vdsshuman, K12human, K21human were set as 45.1, 81.0 mL/kg, 0.275, 0.355 d−1, respectively (Table 1). K10human was approximated by the following equation:
 | (10) |
| Parameters | K10 d−1 | K12 d−1 | K21 d−1 | V1 mL/kg | Vdss mL/kg |
|---|---|---|---|---|---|
| Geometric mean | 0.0832 | 0.275 | 0.355 | 45.1 | 81.0 |
| Geometric CV (%) | 57.8 | 92.9 | 96.1 | 22.6 | 23.8 |
βhuman was calculated by Eq. 9 and Vdsshuman was taken to be 81.0 mL/kg, as above.
To simulate the human plasma concentration–time profile after i.v., the two-compartment model described in Eq. 1 was used.
 | (11) |
 | (12) |
 | (13) |
 | (14) |
The human plasma concentration–time profiles after i.v. dosing were predicted using allometrically scaled NHP parameters in the two-compartment model.13,27) Scaling was conducted following Eq. 9, and scaling exponents were 1.0 and −0.15 for V1 and rate constants (K10, K12, and K21), respectively. To simulate the human plasma concentration–time profile after i.v. dosing, the two-compartment model described in Eqs. 1 and 11–14 was adopted.
Table 1 lists the human geometric mean values from 24 mAbs for two-compartment model parameters. Although, K12 and K21 had large coefficients of variation (CV) with 92.9 and 96.1%, the CV of V1 and Vdss were small at 22.6 and 23.8%, respectively. Therefore, the ratio between K12 and K21 would be constant.
Two-Compartment Model Analysis of Human mAb PK after i.v. AdministrationUsing the two-compartment model parameters for the collected human PK data of 24 mAbs, the PK features in humans were analyzed. The human serum/plasma concentration–time profile consisted of the distribution phase with a short half-life and the elimination phase with a long half-life. In the simulated human PK profile using human geometric means for the two-compartment model parameters, the distribution phase ceased to contribute from 2.24 d after i.v. dosing because the elimination phase accounted for over 80% of serum/plasma concentration from that time. In the dose-normalized PK profile, the differences in serum/plasma concentration within the distribution phase were small, and the difference in PK between mAbs emerged in the elimination phase (Fig. 2A). Raxibacumab shows a human PK profile that is typical of two-compartment model analysis after i.v. (Fig. 2B). AUCinf was mainly covered by AUCβ in human after i.v. dosing, and the contribution percentage to AUCinf exceeded 90% (Fig. 2C). Therefore, CL could be evaluated by AUCβ. Additionally, Vdss showed approximately comparable values with Vdβ, and t1/2β showed an inverse correlation with CL (R2 = 0.824) (Figs. 2D, 2E). These features indicate that prediction of human t1/2β is needed for predicting the human plasma concentration–time profile.

(A) Dose-normalized human PK profile for 24 mAbs analyzed by the two-compartment model (dotted lines) and simulated human PK profile using human geometric mean values (solid line). (B) A typical concentration–time profile found by the two-compartment model (raxibacumab). (C) Fraction percentages of exposure in human for the distribution phase (AUCα) and elimination phase (AUCβ), respectively. (D) Relationship between distribution volume at steady state (Vdss) and distribution volume at elimination phase (Vdβ) in human. Solid diagonal line represents line of unity. The dotted lines represent two-fold range above and below the line of unity. (E) Relationship between human clearance (CL) and half-life at elimination phase (t1/2β) in human. The dotted line is the regression (exponential regression) line.
Distribution volume was analyzed using data from 24 mAbs for humans and 19 mAbs for NHPs. There was no correlation in V1 and Vdss between humans and NHPs (Figs. 3A, 3B) and furthermore, the scatter plots of V1 and Vdss in both species showed the values centralized to a geometric mean; therefore, each species had specific values for the distribution parameters of mAbs, and the geometric mean values (CV%) of Vdss in humans and NHPs were 81.0 mL/kg (23.8) and 71.1 mL/kg (35.3), respectively. The relationship between human K12 and K21 was represented by the equation K21 = 1.32 × K12 (R2 = 0.665) (Fig. 3C), and this correlation indicated that the ratio was constant between V1 and V2. Moreover, these results corresponded to the small CV of Vdss among mAbs.

(A) Human distribution volume of central compartment (V1) is plotted against NHP V1. (B) Human distribution volume at steady state (Vdss) is plotted against NHP Vdss. (C) Relationship between human central to peripheral distribution rate (K12) and human peripheral to central distribution rate (K21). The dotted line is the regression (linear regression) line.
As explained earlier, predicting the human t1/2β was important for predicting the human plasma concentration–time profile, so allometric scaling for predicting human t1/2β from the NHP t1/2β was calculated. Allometric scaling was conducted for NHP data on 19 mAbs, and the scaled values were plotted against human values (Fig. 4). Allometric scaling by 78.9 and 89.5% of the NHP t1/2β gave predictions that were within 2- and 3-fold of actual human t1/2β, respectively. Therefore, human t1/2β could be predicted from NHP t1/2β with allometric scaling.

Predicted values for human t1/2β were plotted against observed values for human t1/2β. Human t1/2β was predicted from NHP data using simple allometry with the exponent of −0.15. When body weight was not reported, it was calculated using 70 kg in human, 6 kg in rhesus monkey, and 3.75 kg in cynomolgus monkey . Solid, dotted, and dashed gray lines indicate 100%, 50–200%, and 33–300% prediction, respectively. Solid diagonal line represents line of unity. The dotted lines represent two-fold range above and below the line of unity. The dashed lines represent three-fold range above and below the line of unity.
To predict the human plasma concentration–time profile after i.v. dosing, we propose the half-life method, which only requires data on t1/2β from NHP (Fig. 5). Based on the above findings on human PK, it was not necessary to extrapolate values from non-clinical data to predict human V1, K12, K21, which mainly contribute to the distribution phase for the human plasma concentration–time profile of mAbs. In the half-life method, human geometric mean values were applied to V1, K12, and K21. Finally, K10 was estimated using allometrically scaled β of NHP and the human geometric means of V1 and Vdss.

NHP, non-human primate; PK, pharmacokinetics; V1, distribution volume of central compartment; K12, central to peripheral distribution rate; K21, peripheral to central distribution rate; β, elimination rate constant at elimination phase; BW, body weight; K10, central elimination rate; Vdss, distribution volume at steady state.
The half-life method was verified for 19 mAbs for which PK data after i.v. dosing both in humans and NHPs were available. With the proposed method, all of the values predicted for human K10 of mAbs were within 2-fold of the predicted value using allometric scaling (Fig. 6). The difference in the parameter values predicted with allometric scaling exceeds 2-fold in about 20% of mAbs (Fig. 4). Therefore, when predicting the human plasma concentration–time profile, the half-life method using only NHP t1/2β successfully obtained estimations comparable to those from a two-compartment model analysis with allometric scaling.

Predicted human K10 by half-life method were plotted against predicted values using allometric scaling method. Solid diagonal line represents line of unity. The dotted lines represent two-fold range above and below the line of unity.
In this study, a novel prediction method (the half-life method), which uses only t1/2β from NHP data, was developed to predict the human plasma concentration–time profile of mAbs after i.v. administration (Fig. 5). The half-life method is based on features of human and NHP PK revealed in this report. Moreover, the results from this new approach were comparable with those from conventional approaches, such as the allometric scaling and Dedrick methods.
According to the data we collected, mAbs exhibit bi-phased elimination decay after i.v. dosing in humans (Fig. 2A), so a two-compartment model analysis was also conducted, giving CV values for human V1 and Vdss of about 20% (Table 1). The margin of error for these CV values is comparable with quantification methods; for example, in guidelines for a ligand-binding assay generally used to quantify mAbs, precision and accuracy are defined as within 20%.64) Additionally, the geometric mean of V1 was comparable to the plasma volume in humans and NHPs.65) These small differences in CV supported the idea that distribution of mAbs would be strictly limited by their large molecular weight and high hydrophilicity. Almost all AUCinf consisted of AUCβ in human, and CL could be evaluated by plasma concentration at the elimination phase (Fig. 2C). The values of Vdβ were similar among mAbs, and t1/2β had inverse correlation to CL (Fig. 2E). Moreover, these PK features were common in humans and NHPs, so CL also showed an inverse correlation to t1/2β in NHPs (data not shown). Generally, human CL is predicted from the CL of NHP by allometric scaling using 0.85 as the scaling exponent; therefore, human t1/2β could be predicted from t1/2β of NHP using the same scaling exponent21) (−0.15 as rate)21) (Fig. 4). These results indicated that to predict the human plasma concentration–time profile, V1, Vdss parameters did not need to be extrapolated from NHP values.
We proposed the half-life method for predicting the human plasma concentration–time profile of mAbs after i.v. dosing based on features revealed in the data collected from literature. A human geometric mean value was applied to predict V1 because the CV value among mAbs was small. Although human K12 and K21 showed large differences between the 24 mAbs, the ratios between K12 and K21 were similar (Fig. 3C). Moreover, the contribution of the distribution phase to AUCinf in humans was small. Therefore, the human geometric means were adopted for K12 and K21. In human PK of mAbs, AUCβ was a main contributor to exposure, and Vdss was approximated as Vdβ (Figs. 2C, 2D), which led to the following approximations: CL = K10 × V1 = β × Vdβ ≈ β × Vdss, and K10 were calculated following Eq. 9. The same relationship was confirmed in NHP, and human β (β = Ln(2)/t1/2β) could be predicted from the t1/2β of NHP. Therefore, human β was predicted by extrapolating from NHP data, using a scaling exponent of −0.15. A human geometric mean value was applied to calculate Vdss because of the small variation among mAbs. Accordingly, human K10 could be predicted from the t1/2β of NHP. The values for predicted human t1/2β were comparable between the half-life method and a conventional method (Fig. 6, Supplementary 2).
The half-life method predicted similar values for human t1/2β compared with two empirical methods commonly used to predict the human plasma concentration–time profile of mAbs: the two-compartment model with allometric scaling and the Dedrick plot.13,27–29) The values for human t1/2β predicted by the half-life method and this two-compartment model are comparable because: 1) distribution parameters showed little variation both in humans and NHPs; 2) AUCβ was the main contributor to AUCinf in both species; and 3) human CL could be predicted from NHP CL. Although there was a less than 2-fold difference in the predicted human K10 for all mAbs, cetuximab, nivolumab, and humicade showed a different PK profile (Supplementary 2). A mAb with a distribution phase of less than a day and another with a phase lasting close to one week could show two different predicted PK profiles, depending on whether the half-life or allometric method was used; this is because the half-life method only considers the elimination phase. Therefore, in vitro tools for evaluating the distribution phase are needed to improve efficiency in the prediction of human plasma concentration–time profiles. A Dedrick plot would predict similar results in humans as the half-life method, but the Dedrick plot is not model-based and could not be expanded to predict mAbs with non-linear elimination. For these reasons, the half-life method is simpler and more useful than conventional approaches. We found that, in the half-life method, a scaling exponent of −0.15 for t1/2β was more accurate than a scaling exponent of −0.25; that is, 84.2 and 73.7% of estimates were within 2-fold of the actual values, respectively (data not shown). Therefore, the scaling exponent for the half-life method was determined as −0.15. Although only one example was tested, a satisfactory result was observed for motavizumab-YTE, a mAb mutated to have longer half-life in plasma, which suggests that the half-life method would be useful for predicting the PK of mutated mAbs.
A feature of EPO and G-CSF is their specific binding to a receptor, which means that receptor-mediated endocytosis contributes to the non-linear CL of these peptides.9,10) Several mAbs exhibited non-linear PK because of the binding to membrane antigen, which is a biological mechanism of action. Therefore, predicting the plasma concentration–time profile of mAbs that have non-linear PK is important not only for PK but also for pharmacology and toxicology. Although several studies have been conducted to predict the PK in humans for mAbs that showed non-linearity, many of the mechanisms of non-linear PK still need to be thoroughly understood.15,22) The models needed to predict non-linear PK in humans consist of non-linear and linear elimination processes.13,27,32) The half-life method can elucidate the human plasma concentration–time profile for linear elimination, which may then contribute to predicting the non-linear elimination of a mAb.
The half-life method can contribute to the principle of Refinement as part of the 3Rs (Refinement, Reduction and Replacement) initiative. Avoiding unnecessary injections and blood draws palliates the pain suffered by animals and is important to animal welfare.5) Unfortunately, blood must be drawn more than 10 times for PK analysis in NHP studies; it is drawn frequently in a short span of days after i.v. administration to evaluate the distribution phase. The conventional prediction method requires evaluating every PK profile after i.v. administration, whereas the half-life method does not need the distribution phase for NHPs, but only the t1/2β (Refinement), and this value can be derived from plasma concentration data with just three time points in the elimination phase. Conventional PK studies utilizing frequent blood draws could be classified as category C according the SCAW system, however a lesser frequency of only three blood draws once a week frequency falls into Category B according to some institutional animal care and use committees (IACUCs).66)
Furthermore, to improve animal welfare, it is also important to reduce the total number of animals used in such experiments. While the half-life method on its own does not fulfill the 3R initiative’s Reduction principle, it only requires NHP t1/2β to predict the human plasma concentration-time profile, and this could already be provided by a related pharmacology or toxicology study.67–69) Therefore, by using the half-life method to avoid an NHP PK study, we may indirectly reduce the number of animals involved in overall mAb development. This study was able to elucidate the features of human PK, and the human profile could be predicted using just the t1/2β obtained from NHP. The half-life method uses just one parameter obtained from NHPs to predict human t1/2β. Therefore, an in vitro assay system which evaluates human t1/2β could replace an NHP study intended for the human plasma concentration–time profile prediction. Single PK parameters such as t1/2 have been evaluated in vitro, but there are no reports of in vitro systems able to evaluate all four parameters in two-compartment model for human PK prediction.70–72) The comparable value of t1/2 can be substituted for t1/2β in cases of mAbs with linear elimination. The combination of the half-life method and the in vitro evaluation of human t1/2 could further improve both efficiency and animal welfare in mAb development. In particular, speed would improve, because the evaluation of in vivo PK takes a long time due to the slow elimination from circulation. Replacing an NHP study performed for human PK profile prediction with an in vitro assay system would also reduce the number of NHPs involved.
In conclusion, to increase the efficiency of mAb development and in consideration for animal welfare, we have developed a half-life method that uses only the t1/2β of NHPs to predict the human plasma concentration-time profile of mAbs. Moreover, the accuracy of this half-life method is comparable to that of conventional methods.
The authors thank Sally Matsuura and Jacob Davis for an advice in the preparation and the language editing of this paper.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.