2021 Volume 44 Issue 11 Pages 1790-1795
2021 Volume 44 Issue 11 Pages 1790-1795
Vibrio vulnificus can utilize the xenosiderophore desferrioxamine B (DFOB) as an iron source under iron-restricted conditions. We previously identified in V. vulnificus that transcription of the desA gene encoding the outer membrane receptor for ferrioxamine B (FOXB) is activated by the AraC-type transcriptional regulator encoded by desR together with DFOB. In this study, we overexpressed and purified DesR as a glutathione S-transferase-fused protein and examined interaction between the promoter region of desA and DesR. Electrophoretic mobility shift assay (EMSA) revealed that DesR directly binds to the regulatory region of desA, and this binding was enhanced by the presence of DFOB in a concentration-dependent manner, while the presence of FOXB did not affect the potentiation of their binding. Moreover, EMSA identified that DNA fragments lacking a probable DesR binding sequence were unable to form complexes with DesR. Finally, deoxyribonuclease I footprinting assay demonstrated that the DNA binding sequence of DesR is located between −27 and −50 nucleotides upstream of the desA transcription start site. These results strongly indicate that DesR can directly activate the transcription of desA in cooperation with DFOB, which acts as a coactivator for DesR.
In almost all bacteria, iron is an essential element for maintaining physiological functions.1) However, the bioavailability of iron is extremely low because it forms insoluble ferric hydroxide complexes under aerobic conditions and at neutral pH.2) Therefore, many bacteria have developed iron acquisition strategies via siderophores (Sids), small ferric iron-chelating molecules produced and secreted by microorganisms, to gain access to iron from such iron-restricted extracellular environments.3) The first step of entry into Gram-negative bacteria of a ferric iron-chelating Sid (Fe3+-Sid) is mediated by active transport involving a specific outer membrane receptor (OMR) together with the energy-transducing TonB-ExbB-ExbD system. The Fe3+-Sid translocated into the periplasm via a specific OMR is then translocated into the cell cytosol through a specific ATP-binding cassette transporter.3) The expression of genes involved in Fe3+-Sid uptake is regulated by the ferric uptake regulator (Fur), which is a ubiquitous global repressor in Gram-negative bacteria.4)
Vibrio vulnificus is a Gram-negative halophilic bacterium ubiquitous in warm coastal waters and is known as a causal agent for gastroenteritis, wound infections, and primary septicemia.5) Patients with liver disease and hemochromatosis, who frequently have elevated iron levels, are predisposed to V. vulnificus infections because they have iron stores for bacterial growth. Furthermore, desferrioxamine B (DFOB), which is a hydroxamate siderophore produced by many Actinomycetes, is often used as an iron-chelating agent for chronic iron overload, and it facilitates the proliferation of some pathogens, including V. vulnificus.6)
V. vulnificus secretes its cognate siderophore, vulnibactin, and transports extracellular iron as ferric vulnibactin via the specific OMR VvuA and unidentified ABC transporter(s) associated with periplasmic binding proteins FatB and VatD.7,8) In addition to producing vulnibactin, V. vulnificus can pirate aerobactin as a xenosiderophore by expressing its cognate OMR IutA and an ABC transporter complex, VatCDB.9,10) In our previous study, it was also shown that this bacterium can also utilize desferrioxamine B as xenosiderophore via the specific OMR DesA and subsequently via VatCDB, and that the expression of desA is transcriptionally regulated not only by the Fur repressor but also by the DesR activator.11) In this report, we show that V. vulnificus DesR binds well to the promoter region of the desA gene in the presence of DFOB, not ferrioxamine B (FOXB, an iron-bound form of DFOB), as a coactivator. Our results strongly suggest that DFOB-associated DesR interacts with the desA promoter region, thereby increasing desA gene expression.
The mesylate salt of DFOB was purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). FOXB was prepared by mixing DFOB and FeCl3 in a 1 : 1 M ratio.
PrimersThe PCR primers used in this study are listed in Table 1.
| Purpose for | Primer name | Sequence (5′–3′)* |
|---|---|---|
| Amplification of desR | VvdesR-NdeI-F | GTATTCCATATGGCAACAGCACAAAGTAGAGAC |
| VvdesR-EcoRI-R | GGTGATGAATTCGAGCTTGGATAACCTCATAC | |
| Construction of probe 1 | VvdesEMSA-F1 | GTACCGATCCAAAAAGTATG |
| flu-VvdesEMSA-R | fluorescein-GTTCATATTGTCCCTAAATACC | |
| Construction of probe 2 | VvdesEMSA-F2 | GACTTTAAGTTTGTGTTGAA |
| flu-VvdesEMSA-R | See above | |
| Construction of probe 3 | VvdesEMSA-F3 | ATCTTTTGAGCTGCCCAATA |
| flu-VvdesEMSA-R | See above | |
| Construction of probe 4 | VvdesEMSA-F1 | See above |
| Vvdesdel-1 | ctcaaaagatttcaacacaaACTTAAAGTC | |
| Vvdesdel-2 | ttgtgttgaaatcttttgagCTGCCCAATA | |
| flu-VvdesEMSA-R | See above | |
| Construction of sense probe | VvdesApro-foot-F2-Tx | Texas Red-CAATGCTGAAATGGAATACTG |
| VvdesApro-foot-R | GGTAAACGTAGCCATGATCG | |
| Construction of antisense probe | VvdesApro-foot-F | GCATTGATGGTTATGAACTGG |
| VvdesApro-foot-R2-Tx | Texas Red-TTAAATGGAGTTGTTGTGTTC |
* Underlined sequences indicate restriction within VvdesR-NdeI-F and VvdesR-EcoRI-R represent NdeI and EcoRI sites, respectively; lowercase letters within Vvdesdel-1 and Vvdesdel-2 represent complementary base pairs.
The coding region of desR was amplified by PCR using chromosomal DNA isolated from V. vulnificus M2799 and a set of primer pairs, VvdesR-NdeI-F and VvdesR-EcoRI-R. The PCR fragment was ligated into the NdeI/EcoRI cloning sites in the pCold-glutathione S-transferase (GST) expression vector (TaKaRa, Shiga, Japan), yielding plasmid pVvDesR.
Overexpression and Purification of GST-DesROverexpression of GST-DesR was carried out using pVvDesR by the same method previously described.10) Purification of GST-DesR was performed with GSTrap FF affinity column (GE Healthcare, Chicago, IL, U.S.A.), according to the method previously described.10) Purified GST-DesR was then concentrated and desalted by buffer exchange with a storage buffer (100 mM Tris–HCl, 50 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 40% glycerol, pH 8.0) and stored at −20 °C.
Electrophoretic Mobility Shift Assay (EMSA) of the desA Regulatory RegionEMSA was performed as previously described.10) For EMSA experiments, fluorescein-labeled DNA probes 1 to 3 were amplified by PCR using V. vulnificus chromosomal DNA as template and the following three sets of primer pairs: probe 1, VvdesEMSA-F1 and flu-VvdesEMSA-R; probe 2, VvdesEMSA-F2 and flu-VvdesEMSA-R; and probe 3, VvdesEMSA-F3 and flu-VvdesEMSA-R. Moreover, the fluorescein-labeled DNA probe 4 containing a deletion of probable DesR binding sequence was prepared by overlap extension PCR12) as previously described13) using the following primers: VvdesEMSA-F1, Vvdesdel-1, Vvdesdel-2, and flu-VvdesEMSA-R. EMSA reaction mixtures (10 µL) contained 0.01% BSA, 0.1 µg/µL poly(dI-dC), 25 nM labeled probe, and varying concentrations of GST-DesR in binding buffer (20 mM Bis-Tris borate, pH 7.5, 50 mM KCl, 1 mM MgCl2, 5% glycerol). As required, DFOB or FOXB was added to the reaction mixture at the indicated concentrations. The reaction mixtures were incubated at 25 °C for 30 min before separation of probe/GST-DesR complexes by electrophoresis on 5% native polyacrylamide gels in 0.5 × TBE buffer at 100 V for 1 h. Gel images were analyzed with a ChemiDoc Touch imaging system (BioRad, Hercules, CA, U.S.A.) using fluorescein excitation/emission filters.
Deoxyribonuclease (DNase) I Footprinting Assay for the desA Regulatory RegionDNase I footprinting assays were performed as previously described.10) Double-stranded DNA probes labeled with Texas Red were prepared by PCR using V. vulnificus chromosomal DNA as template and the following two sets of primer pairs: sense probe, VvdesApro-foot-F2-Tx and VvdesApro-foot-R; antisense probe, VvdesApro-foot-F and VvdesApro-foot-R2-Tx. Varying amounts of GST-DesR ranging from 0 to 20 pmol were incubated with 0.5 pmol Texas Red-labeled probe in a total volume of 40 µL in binding buffer, as described for EMSA. As required, 50 nmol of DFOB was added to the reaction mixture. After incubation for 30 min at 25 °C, 0.1 U DNase I (Promega, Madison, WI, U.S.A.) was added to each reaction mixture, followed by further incubation at 30 °C for 12.5 min. The reaction was stopped by phenol-chloroform extraction followed by ethanol precipitation. The pellets were directly suspended in 2 µL formamide loading dye, denatured for 5 min at 100 °C, and separated using an SQ5500E DNA sequencer (Hitachi High-Tech, Tokyo, Japan) on a 6% polyacrylamide/urea gel along with a DNA sequencing ladder obtained with the same labeled primer as a marker.
In our previous study, we identified in V. vulnificus that the desR gene, a member of the AraC family of transcriptional regulator genes, plays a role in DFOB-inducible desA expression. Moreover, the N-terminal region of DesR exhibits significant similarity to AraC family regulators of Escherichia coli, such as MarA, SoxS, and Rob.11) It has been reported that MarA, SoxS, and Rob regulate the expression of their target genes by binding the 20 bp consensus marbox sequence (AYnGCACnnWnnRYYAAACn) present in their promoter regions.14,15) The promoter region of desA also possesses a probable marbox sequence (GTT GCA CCA AAT TGT TAA CT; conserved nucleotides are underlined),11) but it has not been revealed whether DesR actually binds to the sequence. Therefore, in order to determine whether DesR binds to the regulatory region of desA, EMSA was performed with purified GST-DesR together with a fluorescein-labeled DNA probe (probe 1) that contained the desA regulatory region spanning bp positions −121 to +49 from the desA transcriptional start site (+1) (Fig. 1A). EMSA results indicated that GST-DesR could bind to the probe 1 at 1 µM GST-DesR (Fig. 1B, panel a). In contrast, when DFOB was present in the EMSA reaction mixture, binding between probe 1 and GST-DesR was observed at a lower concentration of GST-DesR than in the absence of DFOB (Fig. 1B, panel b). On the other hand, interestingly, no promotion of binding between probe 1 and GST-DesR was observed in EMSA reaction mixtures containing FOXB in comparison to vehicle only (Fig. 1B, panel c). These results strongly suggested that the binding of DesR to the regulatory region of desA is enhanced by the presence of DFOB, but not FOXB, which functions as a coactivator for DesR. Moreover, we explored the effect of a range of DFOB concentrations on the binding between the regulatory region of desA and DesR (Fig. 1C). As expected, GST-DesR displayed little binding to probe 1 in the absence of DFOB, and the intensity of the probe 1/GST-DesR complex increased when the concentration of DFOB, but not FOXB, increased. Next, to assess whether DesR binds to a probable marbox sequence in the regulatory region of desA, EMSA was carried out with a series of fluorescein-labeled DNA probes (probes 2 to 4) (Fig. 1A). EMSA results showed that GST-DesR could bind to probe 2 (with positions −70 to +49) containing a probable marbox as well as to probe 1 (Fig. 1D). However, GST-DesR was not able to bind to either of probes 3 (positions −30 to +49) or 4 (positions −121 to +49, with positions −50 to −31 deleted) (Fig. 1D). These results indicated that the marbox sequence is required for DesR binding to the desA regulatory region.
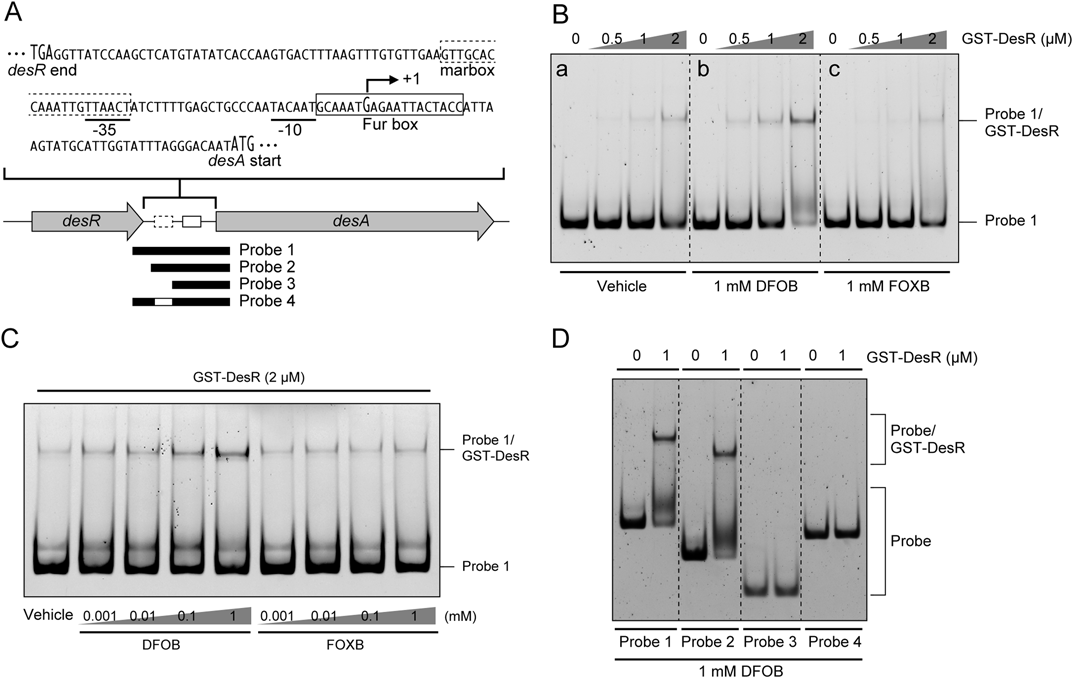
(A) The putative −10 and −35 bp elements are indicated by underlining. The probable marbox and Fur box sequences of the desA promoter are shown as dotted and solid line boxes, respectively. The transcriptional start site (+1) of desA identified in a previous study is indicated by a bent arrow above the nucleotide sequence.11) The closed thick lines indicate the location of fluorescein labeled probes (probes 1 to 4) used for EMSA, and the open line in probe 4 represents the deletion of a probable marbox sequence. (B) EMSA results showed the ability of GST-DesR to bind the desA promoter. EMSA was carried out using 25 nM probe 1 in the presence of GST-DesR at concentrations of 0, 0.5, 1, and 2 µM. As required, H2O vehicle (panel a), 1 mM DFOB (panel b), or 1 mM FOXB (panel c) was added into EMSA reaction mixtures. (C) EMSA results exhibit that DFOB improves the GST-DesR interaction with the desA promoter in a concentration-dependent manner. EMSA was carried out using 25 nM probe 1 in the presence of GST-DesR at a concentration of 2 µM and of DFOB or FOXB at concentrations of 0.001, 0.01, 0.1, and 1 mM. (D) The marbox is required for DNA shifting by GST-DesR. EMSA was performed using 25 nM probes 1 to 4 in the presence of GST-DesR at 0 and 1 µM and of DFOB at 1 mM.
To determine more precisely the DesR-binding sequence in the regulatory region of desA, DNase I footprinting assay was carried out with both 5′ Texas Red-labeled coding and non-coding strand fragments, including a probable marbox in the desA promoter region. Each marbox on the coding and non-coding probes was digested by DNase I in the absence of GST-DesR (Fig. 2, lanes 1, 5, 9, and 13). Moreover, although 5 and 10 pmol of GST-DesR did not affect digestion of the probes by DNase I, 20 pmol of GST-DesR could protect DNA sequences from DNase I digestion, including a probable marbox between bp positions −51 and −27 on the coding probe and between bp positions −53 and −29 on the non-coding probe (Fig. 2, lanes 2 to 4 and 10 to 12). In contrast, in the presence of DFOB, this DesR binding sequence was protected by 10 pmol of GST-DesR, which was less than that in the absence of DFOB (Fig. 2, lanes 6–8, 14–16).
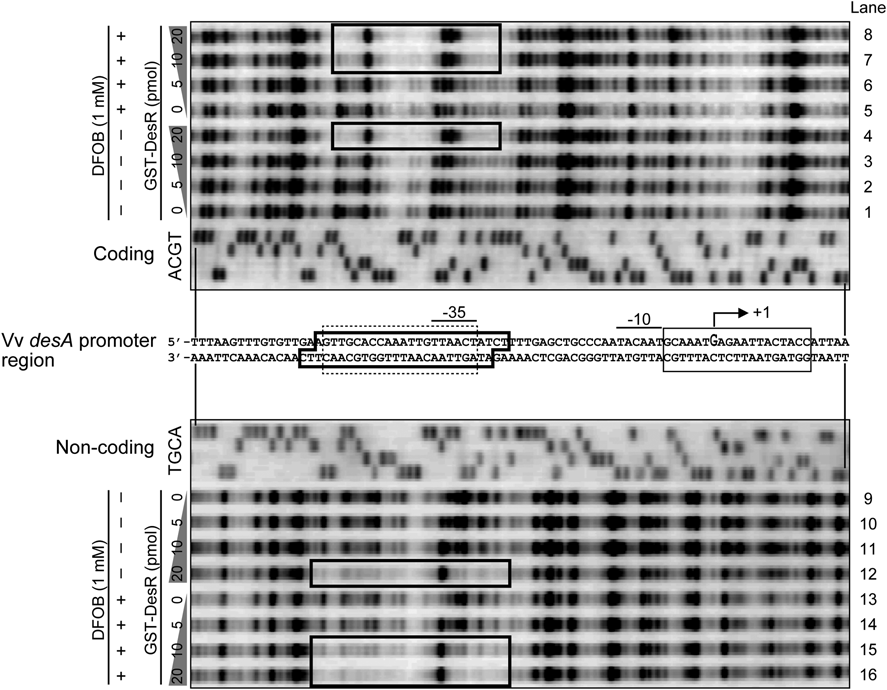
The 5′-ends of the DNA fragments containing the desA promoter region were end-labeled with Texas Red. The presence and absence of DFOB at a concentration of 0 or 1 mM are indicated by the positive and negative signs, respectively. The A, C, G, and T lanes show the results of dideoxy chain termination DNA sequencing reactions. Areas protected by GST-DesR are enclosed by thick lines in the sequence ladders and the nucleotide sequence of the desA promoter region. The putative −10 and −35 elements, marbox, and Fur box in the iutA promoter, are indicated by overlines, dotted line box, and solid line box, respectively. The transcriptional start site (+1) of desA is indicated by a bent arrow above the nucleotide sequence.
In conclusion, we revealed that the DesR-binding sequence in the promoter region of desA contains a probable marbox. As previously reported, DesR showed homology with MarA, SoxS, and Rob in E. coli belonging to the AraC family of transcription factors.11) Marbox is an asymmetric sequence and is classified into two promoter classes according to its orientation and positional relationship with the promoter sequence.14) One is referred to as a class I promoter, and a marbox of this type is located 18 to 19 bp upstream of the −35 bp sequence in the opposite orientation. Accordingly, an AraC family of transcription factor interacts the RNA polymerase (RNAP) α subunit C-terminal domain (αCTD) to activate transcription.14) The other is termed a class II promoter, and a marbox of this form exists in a forward overlap with the −35 bp sequence to be involved in a contact with region 4 of the RNAP σ subunit and αCTD.16) DNase I footprinting examined in this study clarified that the marbox, including the DesR binding sequence, belongs to the class II promoter type (Fig. 2). In addition, we also found that DFOB facilitates binding between the DesR-binding sequence and DesR. DesR, like Rob, is a large protein consisting of two domains. DesR and Rob share identity at the N-terminal, with a helix-turn-helix DNA-binding domain involved in the activation of transcription upon binding to marbox of the respective promoter region.11,17) The C-terminal domain of DesR does not show similarity to that of Rob, because this domain of Rob represents the site for recognizing its unique coactivators, such as bile salts and dipyridyl.18,19) Therefore, it may be hypothesized that the C-terminal domain of DesR should bind to DFOB, thereby enabling DesR to activate transcription of desA.
A proposed induction mechanism for DesA expression is shown in Fig. 3. Under iron-restricted conditions, desR and desA genes of V. vulnificus would be co-transcribed as a polycistronic mRNA like those of V. furnissii,20) but the amount of desA transcript at this time is very low, and thus DesA is hardly produced.11,20) However, when DFOB is present in the environment, it is taken up as FOXB into V. vulnificus cells via DesA, which is lowly expressed under iron-restricted conditions.11,20) FOXB taken into V. vulnificus cells should be reduced by a mechanism that is not yet well understood, leading to the release of ferrous iron and the formation of DFOB. DesR then interacts with DFOB to become active, and its active form interacts with the DesR binding region of the desA promoter and activates desA transcription to increase the production of DesA. In general, when ferric hydroxamate-type siderophores are transported into the bacterial cytoplasm, they are converted in a short time into iron-free siderophores and ferrous ion by ferric siderophore reductases.21–23) Therefore, it seems reasonable that DesR interacts with DFOB rather than FOXB. It is known that DFOB is mainly produced by Actinomycetes,24) a group of bacteria found ubiquitously in soil as well as in seawater and marine sediments.25,26) This mechanism for sensing and pirating DFOB should be highly advantageous to this species because they compete with other neighboring microbes for survival and proliferation in various environmental niches.27) Future studies, including crystal structure analyses, will be focused on understanding how DesR interacts with the DesR binding region of the desA promoter as well as its coactivator molecule, DFOB.
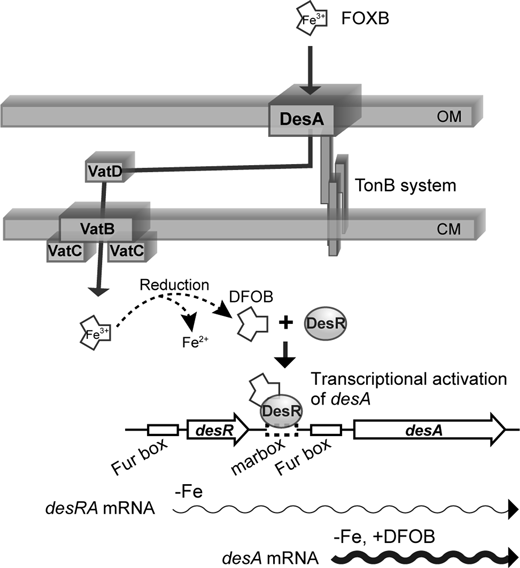
In iron-restricted condition, Fur inactivated by the loss of an iron co-repressor should be released from Fur binding sites (Fur boxes) in the promoter regions of desR and desA.11,20) At this time, desA is probably co-transcribed with desR (desRA mRNA; thin wavy arrow)20); however, as DesR alone can barely bind to marbox in the desA promoter region, desA is rarely transcribed. In contrast, DesR interacts with DFOB, which is taken up as FOXB from the surrounding environment and then reduced, and thereby binds to the region including the marbox in the desA promoter region. As a result, the desA promoter becomes fully active, and desA is abundantly transcribed (thick wavy arrow).
The authors declare no conflict of interest.