2021 年 44 巻 4 号 p. 557-563
2021 年 44 巻 4 号 p. 557-563
Elevated expression of β4-galactosyltransferase (β4GalT) 3 is correlated with poor clinical outcome of neuroblastoma patients. Our recent study has revealed that the transcription of the β4GalT3 gene is activated by Specificity protein (Sp) 3 in SH-SY5Y human neuroblastoma cell line. Here we report the biological significance of the Sp3 phosphorylation in the transcriptional activation of the β4GalT3 gene. The treatment of SH-SY5Y cells with 10% fetal bovine serum (FBS) increased the mitogen-activated protein kinase (MAPK) signaling and the promoter activity of the β4GalT3 gene. Meanwhile, the treatment with U0126, an inhibitor for MAPK kinase, decreased the MAPK signaling and the promoter activity. These findings indicate that the transcriptional activation of the β4GalT3 gene is mediated by the MAPK signaling. In SH-SY5Y cells cultured in the medium containing 10% FBS, the serine (Ser) residues in Sp3 were phosphorylated. Human Sp3 contains four Ser residues, Ser73, Ser563, Ser566, and Ser646, as the putative phosphorylation sites. Sp3 mutant with the mutation of Ser73 did not decrease the promoter activation of the β4GalT3 gene, indicating that Ser73 is uninvolved in the promoter activation of the β4GalT3 gene by Sp3. In contrast, Sp3 mutants with the mutations of Ser563, Ser566, and Ser646 significantly reduced the promoter activation by Sp3. The results suggest that the phosphorylation of these Ser residues is implicated in the promoter activation by Sp3. This study demonstrates that the phosphorylation of Sp3 plays important roles in the transcriptional activation of the β4GalT3 gene in human neuroblastoma.
Neuroblastoma is a common pediatric malignant solid tumor that is derived from neural crest cells of sympathetic nervous system.1,2) Although the overall prognosis of patients with neuroblastoma has remarkably improved by recent advancement of therapies, high risk neuroblastoma is still considered to be one of the most difficult tumors to cure.3) Therefore, the effective therapeutic strategies and drugs for neuroblastoma are required to be developed.
Glycans attached to proteins and lipids are composed of several monosaccharides such as N-acetylgalactosamine, N-acetylglucosamine, galactose, glucose, and mannose. Usually, drastic changes in the glycosylation of cell surface glycoproteins including cell adhesion molecules occur upon malignant transformation of cells.4,5) Accordingly, the functions of cell adhesion molecules are altered in malignantly transformed cells, which is implicated in the malignant potentials of cancer cells.5,6)
β4-Galactosyltransferase (β4GalT) catalyzes the transfer of galactose towards the glycans, and the β4GalT family consists of seven members with different substrate specificities.7,8) Among seven members, β4GalT3 possesses the activities to galactosylate N-glycans terminating with N-acetylglucosamine and lacto-N-triaosylceramide, and synthesize poly-N-acetyllactosamine.9–12) The galactosylated N-glycans synthesized by β4GalT3 are important for the tumor growth, cell migration, and invasion of neuroblastoma cells, and the elevated expression of β4GalT3 is correlated with poor prognosis of patients with neuroblastoma.13) Our recent study has revealed that the transcription of the β4GalT3 gene is regulated by Specificity protein (Sp) 3 in SH-SY5Y human neuroblastoma cell line.14)
Sp3 is the member of Sp family transcription factors that bind to the GC box.15) The Sp3 gene is expressed in most tissues,16) and the gene expression increases in a variety of human cancers including breast, nasopharyngeal, and liver cancers.17–19) Sp3 functions as a transcriptional activator or as a transcriptional repressor depending on the cellular context.15) In our recent study, the β4GalT3 gene promoter has been shown to be activated by Sp3 in SH-SY5Y cells.14) To develop the novel therapeutic strategies and drugs for neuroblastoma, it is necessary to further elucidate the molecular mechanism underlying the promoter activation of the β4GalT3 gene by Sp3 in neuroblastoma.
Sp3 is subjected to post-translational modifications such as acetylation, O-glycosylation, phosphorylation, and sumoylation.15) Among these modifications, the acetylated or the phosphorylated Sp3 functions as a transcriptional activator,20,21) whereas the O-glycosylated or the sumoylated Sp3 functions as a transcriptional repressor.22–24) In the present study, the molecular mechanism underlying the transcriptional activation of the β4GalT3 gene by Sp3 in SH-SY5Y cells was investigated by focusing on the acetylation and the phosphorylation of Sp3.
Anti-Sp3 antibody was purchased from GeneTex, Inc. (Irvine, CA, U.S.A.). Anti-p44/p42 mitogen-activated protein kinase (MAPK) antibody, anti-phospho-p44/p42 MAPK (T202Y204) antibody, and U0126 were from Cell Signaling Technology, Inc. (Danvers, MA, U.S.A.). Anti-phosphoserine antibody was from Sigma-Aldrich (St. Louis, MO, U.S.A.).
Cell CultureSH-SY5Y cells were grown in a mixture of Dulbecco’s modified Eagle’s medium and Ham’s F-12 medium (1 : 1 (v/v)) containing 10% fetal bovine serum (FBS), 50 µg/mL kanamycin, and 1.0 mg/mL glucose.
Preparation of Reporter PlasmidThe reporter plasmid pGL(−2190/+89) containing the 5′-flanking region of the human β4GalT3 gene was prepared using pGL3-Basic vector (Promega, Madison, WI, U.S.A.), which contains the firefly luciferase gene, as described previously.14)
Immunoblotting and ImmunoprecipitationImmunoblotting and immunoprecipitation were performed by the methods described previously.25–27) The extracts of SH-SY5Y cells cultured in the medium containing 10% FBS were immunoprecipitated using anti-Sp3 antibody and μMACS Protein G MicroBeads (Miltenyi Biotec Inc., Auburn, CA, U.S.A.) as per the manufacturer’s instructions. The immunoprecipitated samples were subjected to immunoblotting using anti-Sp3 and anti-phosphoserine antibodies. The band intensity of MAPK and phosphorylated MAPK was determined by analyzing with ImageJ (version 1.52a).26,27)
Treatment with U0126The cells (2 × 105) were transiently transfected with pGL(−2190/+89). U0126 (20 µM) was added to the cells 1 h after transfection with the reporter plasmid.
Preparation of Expression Plasmids for Sp3 MutantsPreviously, pBA-Sp3 was prepared by insertion of the human Sp3 gene into pBApo-EF1α Pur DNA (TaKaRa Bio Inc., Shiga, Japan) as Sp3-expression plasmid.14) As illustrated in Fig. 1, human Sp3 contains one lysine (Lys) residue at amino acid 551 (Lys551) to be acetylated and four serine (Ser) residues at amino acids 73, 563, 566, and 646 (Ser73, Ser563, Ser566, and Ser646) to be phosphorylated (https://www.uniprot.org/uniprot/Q02447).15) In order to substitute these amino acids to alanine (Ala) based on splicing overlap extension-PCR technique,28,29) the mutagenic oligonucleotide primers were designed as listed in Table 1. The positions of the primers and the amplified regions by PCR are shown in Fig. 2. As a representative, the procedure for the preparation of expression plasmid Sp3SM1 was described below. The primers TS28-19 and TS1-26 contained the BamHI and XbaI sites, respectively, and the primers TS1-24 and TS1-25 contained the mutations, which corresponded to the substitution of Ser73 for Ala in the phosphorylation site. First, two overlapped DNA fragments (1–229) and (204–827) containing the mutations were amplified by PCR with pBA-Sp3 as a template and the primer sets (TS28-19 and TS1-25) and (TS1-24 and TS1-26). The PCR condition was as follows: 97 °C, 1.5 min [94 °C, 15 s; 60 °C, 1 min; and 74 °C, 1 min] × 25, and 72 °C, 5 min. Next, the DNA fragment (1–827) containing the mutations corresponding to Ala73 was amplified by PCR with two DNA fragments (1–229) and (204–827) as templates and the primer set (TS28-19 and TS1-26). The PCR condition was as follows: 97 °C, 1.5 min [94 °C, 15 s; 60 °C, 1 min; and 74 °C, 1 min] × 30, and 72 °C, 5 min. The nucleotide sequence of the resultant fragment was verified by DNA sequencing. The DNA fragment (1–827) was exchanged with the corresponding fragment of pBA-Sp3 using the restriction enzyme sites as shown in Fig. 2, to generate the expression plasmid Sp3SM1. Similarly, in order to prepare the other expression plasmids for Sp3 mutants, the DNA fragments containing the mutations were amplified by two round PCR, and then exchanged with the corresponding fragments of pBA-Sp3 using the restriction enzyme sites. The expression plasmids Sp3SM2 and Sp3SM3 were generated for Sp3 mutants containing the substitutions of Ser563 and Ser566, and Ser646 to Ala, respectively. The expression plasmid Sp3SM123 was generated for Sp3 mutant containing the substitutions of all four Ser residues to Ala. Furthermore, the expression plasmid Sp3KM was generated for Sp3 mutant containing the substitution of Lys551 to Ala.
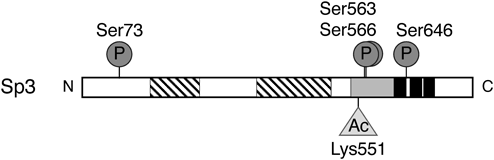
Human Sp3 contains one acetylation site (Lys551) and four phosphorylation sites (Ser73, Ser563, Ser566, and Ser646). Hatched, gray, and black boxes represent the activation domains, the inhibitory domain, and the DNA binding domain that consists of three zinc fingers, respectively. Ac and P indicate the acetylation and the phosphorylation sites, respectively.

Black and white arrows indicate the oligonucleotide primers used, and the white arrows indicate the oligonucleotide primers containing the mutations. The nucleotide sequences of the primers are shown in Table 1.
| Name | Position | Sequence |
|---|---|---|
| TS1-16 | 815/837 | TCGATCTAGATTCTTTGGGACTCTC (XbaI) |
| TS1-17 | 1819/1842 | GATTGGTACCTCTTCCACCACCTTC (KpnI) |
| TS1-18 | 1674/1698 | GTTCTTCTTCCGCGATCCTAATATC |
| TS1-19 | 1639/1663 | GATATTAGGATCGCGGAAGAAGAAC |
| TS1-24 | 204/229 | GATAGGGCCGCCAGCGCCGGGCGAC |
| TS1-25 | 204/229 | GTCGCCCGGCGCTGGCGGCCCTATC |
| TS1-26 | 803/827 | GAATCTAGATCGACACTATTGATTG (XbaI) |
| TS1-27 | 1685/1709 | TCGCTGGTGATGCTACCTTGAATAC |
| TS1-28 | 1674/1698 | AGCATCACCAGCGAGCTGCCACTCT |
| TS1-29 | 1830/1854 | AAGAGGTACCAATCTTGGGAAAAAG (KpnI) |
| TS1-30 | 1924/1948 | CTGCGTTGGCATGCTGGAGAACGCC |
| TS1-31 | 1924/1948 | GGCGTTCTCCAGCATGCCAACGCAG |
| TS28-19 | 1/22 | CGGGATCCATGACCGCTCCCGAAAAGCCCG (BamHI) |
| TS28-20 | 2325/2346 | CCAAGCTTTTACTCCATTGTCTCATTTCCA (HindIII) |
The nucleotide positions are shown from the initiation codon. The cleavage sites of restriction enzymes in parentheses are underlined. The mutations are indicated with bold face letters.
The promoter activity was measured by luciferase assay as described previously.14,25,26) The luciferase assay was conducted in triplicate, and the data represent the mean values with standard deviations of three independent experiments. Statistical significance was evaluated using one-way ANOVA with Tukey’s multiple comparison.
Sp3 is known to be subjected to post-translational modifications.15) Since the acetylated Sp3 functions as a transcriptional activator in MCF-7L human breast cancer cell line,20) the acetylated Sp3 was considered to be implicated in the promoter activation of the β4GalT3 gene in SH-SY5Y cells. As Lys551 in Sp3 has been shown to be acetylated,30) the expression plasmid Sp3KM for Sp3 mutant having Ala instead of Lys was generated. Such substitution was expected to decrease the promoter activity of the β4GalT3 gene due to the lack of the Lys residue to be acetylated. As shown in Fig. 3, the co-transfection of pGL(−2190/+89) with Sp3 increased the promoter activity by 1.8-fold when compared to the co-transfection with pBA (p < 0.05 vs. pBA). Similarly, the co-transfection of pGL(−2190/+89) with Sp3KM enhanced the activity by 2.4-fold without statistically significant differences (Fig. 3), indicating that the promoter activation is observed even though lacking Lys551. These results imply that the acetylation of Lys551 is uninvolved in the transcriptional activation of the β4GalT3 gene by Sp3 in SH-SY5Y cells.
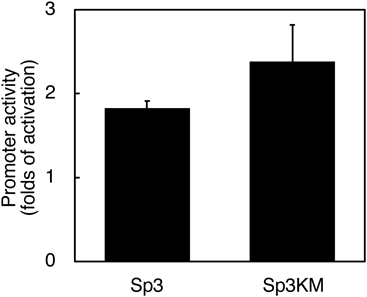
SH-SY5Y cells were transfected with pGL(−2190/+89) and either pBA or expression plasmid (Sp3 or Sp3KM). The promoter activity was expressed as fold activation relative to cells co-transfected with pBA. The data represent the mean values with standard deviations of three independent experiments.
The phosphorylated Sp3 activated the transcription of the vascular endothelial growth factor (VEGF) gene,21) and the Toll-like receptor 5 gene,31) suggesting that the phosphorylation of Sp3 is implicated in the promoter activation of the β4GalT3 gene in SH-SY5Y cells. The phosphorylation of Sp3 has been shown to be mediated by extracellular signaling-regulated kinase (ERK)-MAPK.21,31) Since FBS stimulates the phosphorylation of p44/p42 MAPK through the epidermal growth factor receptor-MAPK signaling,32) we examined whether increasing concentration of FBS stimulates the phosphorylation of p44/p42 MAPK in SH-SY5Y cells. The viability of the 0.5 and 10% FBS-treated cells showed 98% as assessed by trypan blue exclusion. The results showed that the phosphorylation of p44 MAPK increases by 2.1-fold in the medium containing 10% FBS (Fig. 4A), indicating that FBS induces the activation of the MAPK signaling in SH-SY5Y cells. Under the condition, the promoter activity of pGL(−2190/+89) increased by 4.1-fold when compared to 0.5% FBS-treated cells (Fig. 4B). These results suggest that FBS activates the MAPK signaling and leads to the promoter activation of the β4GalT3 gene in SH-SY5Y cells.

A, Immunoblotting with the extracts of SH-SY5Y cells treated with 0.5 and 10% FBS. The blots were probed with the antibodies against p44/p42 MAPK and phospho-p44/p42 MAPK. The band intensity of phosphorylated MAPK against MAPK is indicated below the blots. B, Luciferase assay of the extracts of SH-SY5Y cells transfected with pGL(−2190/+89), followed by the treatment with 0.5 and 10% FBS. The promoter activity was expressed as fold activation relative to cells treated with 0.5% FBS. The data represent the mean values with standard deviations of three independent experiments.
To confirm the involvement of the MAPK signaling in the promoter activation of the β4GalT3 gene, an inhibitor for MAPK kinase, U0126,33) was included in the medium. The viability of the U0126-untreated and treated cells showed 98 and 97%, respectively, as assessed by trypan blue exclusion. Upon treatment with U0126, the phosphorylation of p44 and p42 MAPK decreased to 31 and 10%, respectively, compared with the control (Fig. 5A), indicating that U0126 inhibits the MAPK signaling in SH-SY5Y cells. Under the condition, the promoter activity of pGL(−2190/+89) decreased to 23% when compared to the control (Fig. 5B). Collectively, these findings suggest that the promoter activation of the β4GalT3 gene is mediated by the MAPK signaling in SH-SY5Y cells.

A, Immunoblotting with the extracts of SH-SY5Y cells treated with or without U0126. The blots were probed with the antibodies as described in Fig. 4A. The band intensity of phosphorylated MAPK against MAPK is indicated below the blots. B, Luciferase assay of the extracts of SH-SY5Y cells transfected with pGL(−2190/+89), followed by the treatment with or without U0126. The promoter activity of control was set at 100%. The data represent the mean values with standard deviations of three independent experiments.
Sp3 contains the Ser residues to be phosphorylated, and the MAPK signaling has been shown to increase the Ser phosphorylation of Sp3 in HT29 human intestinal epithelial cell line.31) To analyze whether the Ser residues in Sp3 are phosphorylated in SH-SY5Y cells cultured in the medium containing 10% FBS, we conducted immunoprecipitation using anti-Sp3 antibody followed by immunoblotting with anti-phosphoserine antibody. The results showed that immunoprecipitated Sp3 reacts with anti-phosphoserine antibody (Fig. 6), indicating that the Ser residues in Sp3 are phosphorylated in SH-SY5Y cells. Since the Ser phosphorylation of Sp3 increases in response to the MAPK signaling,31) the Ser phosphorylation of Sp3 is considered to be important for the promoter activation of the β4GalT3 gene in SH-SY5Y cells.
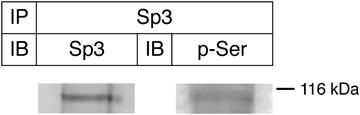
The extracts of SH-SY5Y cells cultured in the medium containing 10% FBS were subjected to immunoprecipitation (IP) using anti-Sp3 antibody. For immunoblotting (IB), the blots were probed with the antibodies against Sp3 and phosphoserine.
As the putative phosphorylation sites, four Ser residues (Ser73, Ser563, Ser566, and Ser646) were reported so far.15) Among these Ser residues, Ser563 and Ser566 are included in the inhibitory domain, and Ser646 is included in the zinc finger DNA binding domain (Fig. 1). In order to investigate the molecular mechanism underlying the promoter activation of the β4GalT3 gene by Sp3, we focused on the roles of the phosphorylation sites in Sp3, and made four expression plasmids for Sp3 mutants, Sp3SM1, Sp3SM2, Sp3SM3, and Sp3SM123, in which the Ser residues were substituted to Ala that cannot be phosphorylated. Since we expected that the roles of Ser563 and Ser566 in the inhibitory domain of Sp3 are similar due to the close location each other, the expression plasmid Sp3SM2 for Sp3 mutant containing the substitutions of both Ser residues to Ala was generated. As shown in Fig. 7, the co-transfection of pGL(−2190/+89) with Sp3 increased the promoter activity by 2.4-fold when compared to the co-transfection with pBA. Similarly, the co-transfection of pGL(−2190/+89) with Sp3SM1 enhanced the promoter activity by 2.4-fold (Fig. 7). The results indicate that the promoter activation is observed even though lacking Ser73, and Ser73 is uninvolved in the promoter activation of the β4GalT3 gene by Sp3. On the contrary, the co-transfection of pGL(−2190/+89) with Sp3SM2 and Sp3SM3 increased the promoter activity by 1.5- and 1.6-fold, respectively (Fig. 7), indicating that the promoter activation by Sp3 mutants decreases significantly by the substitution of Ser563, Ser566, and Ser646 to Ala when compared to the co-transfection with Sp3. When all four Ser residues were substituted to Ala, the co-transfection of pGL(−2190/+89) with Sp3SM123 increased the promoter activity by only 1.2-fold, which exhibited almost similar activity of the pBA-co-transfected cells (Fig. 7). This indicates that the promoter activation by Sp3 is dramatically reduced by the substitutions of all four Ser residues to Ala when compared to the co-transfection with Sp3. Taken together, these results suggest that the phosphorylation of the Ser residues in the inhibitory domain and the DNA binding domain in Sp3 is implicated in the promoter activation of the β4GalT3 gene by Sp3 in SH-SY5Y cells.

A, Schematic drawing of the expression plasmids for Sp3 and Sp3 mutants containing the substitutions of the Ser residues to Ala. B, Luciferase assay of the extracts of SH-SY5Y cells transfected with pGL(−2190/+89) and either pBA or expression plasmid (Sp3, Sp3SM1, Sp3SM2, Sp3SM3 or Sp3SM123). The promoter activity was expressed as fold activation relative to cells co-transfected with pBA. The data represent the mean values with standard deviations of three independent experiments. * p < 0.05, ** p < 0.01.
Recent advancement in development of drugs and biotechnology has become able to overcome the difficulties in cancer therapies. However, neuroblastoma with high risk still shows poor prognosis.1–3) Therefore, it is necessary to develop the effective therapeutic strategies and drugs for neuroblastoma. As the tumor growth, cell migration, and invasion of SK-N-DZ neuroblastoma cell line were suppressed by silencing of β4GalT3 via RNA interference,13) the reagents that can regulate the expression of the β4GalT3 gene are considered to be promising candidates for therapy. In our recent study, the analysis of the transcriptional mechanism has revealed that the transcription of the β4GalT3 gene is regulated by Sp3, and Sp3 activates the β4GalT3 gene promoter in SH-SY5Y cells.14) The understanding of the detailed molecular mechanism underlying the transcriptional activation of the β4GalT3 gene by Sp3 will shed light on the development of the effective therapeutic strategies and drugs for neuroblastoma.
Protein phosphorylation regulates the protein stability, cellular localization, protein–protein interactions, and DNA binding of transcription factors.34) Herein, we demonstrated that the Ser phosphorylation of Sp3, which is mediated probably by the MAPK signaling, is crucial for the transcriptional activation of the β4GalT3 gene in SH-SY5Y cells. Among four Ser residues to be phosphorylated,15) the putative phosphorylation sites Ser563, Ser566, and Ser646 were important for the promoter activation of the β4GalT3 gene by Sp3 in SH-SY5Y cells. As shown in Fig. 1, Ser563 and Ser566 are included in the inhibitory domain, while Ser646 is included in the DNA binding domain of Sp3. This is the first report showing the roles of the phosphorylation sites in the inhibitory domain and the DNA binding domain of Sp3. Although Sp1 and Sp3 belong to the same family, their molecular structures are different partially.15) The inhibitory domain of Sp1 is located at amino acids 2–82 near N-terminus (https://www.uniprot.org/uniprot/P08047), while that of Sp3 is located at amino acids 534–620 between the activation domain and the DNA binding domain (https://www.uniprot.org/uniprot/Q02447). The phosphorylation of Sp1 was implicated in the transcriptional activation and repression, which is probably due to the location of the phosphorylation sites in Sp1.35) The phosphorylation of Ser59 in the inhibitory domain of Sp1 increased the DNA binding ability to the dihydrofolate reductase gene promoter, and activated the transcription.36) Similarly, our results demonstrated that the phosphorylation of Ser563 and Ser566 in the inhibitory domain of Sp3 functions to activate the β4GalT3 gene promoter. From these results, the phosphorylation in the inhibitory domain may function positively in the transcription despite the different molecular structures between Sp1 and Sp3.
Concerning the relationship between the phosphorylation and the DNA binding ability, the increment in the Sp1 phosphorylation by okadaic acid-treatment resulted in the increased DNA binding towards the target DNA.37) In the DNA binding domain of Sp1, the phosphorylation of three sites (Thr668, Ser670, and Thr681) increased the DNA binding ability to the platelet-derived growth factor-D gene promoter, thereby enhancing the promoter activity in human smooth muscle cells.38) Furthermore, in the zinc finger DNA binding domain of GATA4, the phosphorylation of Ser261 was implicated in the DNA binding towards the natriuretic peptide A gene promoter, and the mutation of the phosphorylation site resulted in the impaired promoter activation.39) Therefore, it is likely that the phosphorylation of Ser646 in the DNA binding domain of Sp3 increases the DNA binding, resulting in the promoter activation of the β4GalT3 gene in SH-SY5Y cells.
Ser73 in Sp3 has been shown to be phosphorylated by p42 MAPK in vitro and by Erk in vivo, and the phosphorylation of Ser73 contributed to the transcriptional activation of the VEGF gene promoter without influencing DNA binding ability.21) On the contrary, our study revealed that the phosphorylation of Ser73 is uninvolved in the transcriptional activation of the β4GalT3 gene by Sp3 in SH-SY5Y cells. However, we could not exclude the possibility that Ser73 is not phosphorylated in SH-SY5Y cells, which may be caused by the different cell types.
Interestingly, prominent decrease in the promoter activation of the β4GalT3 gene was observed by co-transfection with Sp3 mutant Sp3SM123, suggesting that the phosphorylation of Ser563, Ser566, and Ser646 functions cooperatively to increase the Sp3 ability to activate the β4GalT3 gene promoter. The possible mechanism is that an intramolecular interaction of the inhibitory domain with the adjacent DNA binding domain is induced by the phosphorylation of both domains and increases to recruit transcriptional cofactors for transcription.
Besides the phosphorylation, the Ser residues are possibly subjected to O-glycosylation,40,41) and a balance between the phosphorylation and the O-glycosylation of Sp1 regulates the transcription of the calmodulin gene.42) Although so far the O-glycosylation of Sp3 occurred in N-terminal portion (amino acids 1–490), and the O-glycosylated Sp3 functioned as a transcriptional repressor,22) the precise O-glycosylation sites remain unknown. If the O-glycosylation of Sp3 is implicated in the transcriptional activation of the β4GalT3 gene, it is necessary to examine whether the O-glycosylation sites share the phosphorylation sites, which were analyzed in this study.
In Sp3, the acetylation is another modification that functions as the transcriptional activator. The acetylated Sp3 activated the transforming growth factor-β receptor type II gene promoter in MCF-7L human breast cancer cell line.20) However, the acetylation is considered to be uninvolved in the transcriptional activation of the β4GalT3 gene by Sp3 in SH-SY5Y cells because the promoter activity increased by Sp3 mutant with the mutation of Lys551. Previously, similar promoter activation by Sp3 mutant with the mutation of Lys551 was observed.30) Since Lys551 is included in the inhibitory domain of Sp3, the acetylation of Lys551 is considered to be necessary for the inhibitory function of Sp3. The lack of Lys551 in Sp3 mutant may attenuate the inhibitory function, thereby increasing the promoter activity of the β4GalT3 gene by Sp3 mutant in SH-SY5Y cells. Alternatively, Lys551 in Sp3 is subjected to sumoylation.23) In general, sumoylation is implicated in the diverse regulatory functions such as protein stability, chromatin structure regulation, subcellular compartmentalization, and transcription factor activity.43,44) The sumoylation usually exhibits the negative effects on the activities of transcription factors.44) The lack of the Lys residue to be sumoylation may also increase the Sp3 ability, which leads to the increased promoter activity of the β4GalT3 gene.
In our recent study, Sp3 has been shown to function as a transcriptional activator for the β4GalT3 gene in SH-SY5Y cells but function as a transcriptional repressor in A549 human lung cancer cell line.14) There are reports that in SH-SY5Y cells Sp3 functions as a transcriptional activator for the genes encoding nicotinamide adenine dinucleotide phosphate oxidase subunit 4 and zipper protein kinase.45,46) Our preliminary study showed that Sp3 functions as a transcriptional repressor for the β4GalT3 gene in SW480 human colon and MKN1 human gastric cancer cell lines like in A549 cells (unpublished data). These observations suggest that Sp3 functions as a transcriptional activator in neuroblastoma but as a transcriptional repressor in other cancer types. From this study, such opposite functions of Sp3 may ascribe to the different phosphorylation state and sites of Sp3 among cancer types. Previously, the phosphorylation of zinc finger homeodomain enhancer-binding protein showed in cell-specific manner,47) and the phosphorylation affected its DNA binding ability and changed the transcriptional activity.48) Therefore, it is of interest to investigate whether the phosphorylation state and sites are different between SH-SY5Y and A549 cells by analyzing the purified Sp3 from both cell lines.
In summary, this study presents a novel insight into the transcriptional activation of the β4GalT3 gene by Sp3 in neuroblastoma that is mediated through the Ser phosphorylation of Sp3 probably by the MAPK signaling. By regulating the phosphorylation state of Sp3, the expression of the β4GalT3 gene can be down-regulated in neuroblastoma, which may suppress the malignant phenotypes of neuroblastoma.
This work was supported by Presidential Research Grant from Nagaoka University of Technology and Japan Society for the Promotion of Science KAKENHI Grant Number 15K07924 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to TS.
The authors declare no conflict of interest.