2022 Volume 45 Issue 9 Pages 1283-1290
2022 Volume 45 Issue 9 Pages 1283-1290
Ubiquitin-specific peptidase 9X (USP9X) has been reported to be closely associated with the formation and progression of a variety of malignant tumors. However, the mechanism by which USP9X is involved in osteosarcoma and development has not been clearly studied. This work aimed to probe the influence of USP9X on osteosarcoma cell proliferation, migration and invasion. This study recruited sixty-seven patients with histologically definited osteosarcoma. Osteosarcoma samples and cell-line were used to reflect the expression level of USP9X. Analysis of cell proliferation by thiazolium blue (MTT) assays. Transwell experiments and wound healing were used to verify cell migration and invasion capabilities. The effect of USP9X was investigated through in vivo experiments. USP9X-related pathway proteins were detected by Western blot and quantitative real-time PCR (qRT-PCR). The expression of USP9X in osteosarcoma was higher than that in adjacent tissues. The overall survival of patients with USP9X-negative patients was better than that of patients with USP9X-positive. The growth of osteosarcoma cells in vivo and in vitro was inhibited by USP9X inhibitor. Cell migration and invasion were significantly inhibited by down-regulation of USP9X. USP9X was involved in extracellular signal-regulated kinase 1/2 (ERK1/2) and phosphatidylinositol-3-kinases/protein-serine-threonine kinase (PI3K/Akt) pathway in osteosarcoma cells. Proliferation, migration and invasion of osteosarcoma cells were inhibited by down-regulation of USP9X, and were related to the ERK1/2 and PI3K/Akt signaling pathways, therefore, it might probably become a new target for the prevention and treatment of osteosarcoma.
Osteosarcoma is a malignant tumor originated from mesenchyma that mainly occurs in children, adolescents, and young adults.1,2) Although modern combination therapies have significantly improved tumor resection and long-term prognosis, 25–35% of patients are initially metastasis-free and subsequently metastatic. These phenomenon remains the leading cause of death.3,4) At the same time, osteosarcoma responds poorly to chemotherapy and has been shown to have a worse prognosis.5,6) Based on the tolerance of drugs in patients with osteosarcoma, especially in advanced patients, we need to broaden our horizons and look for new targeted therapy regimens and effective targets. Ubiquitylation is one of the main types of intracellular protein covalent modification.7,8) It is involved in cell proliferation,9,10) metastasis,11,12) differentiation,13) apoptosis,14) gene expression,15) transcriptional regulation,16) signal transduction,17) injury repair,18) inflammation,19,20) immunity,13,21) and almost all life activities. Current studies have confirmed a large family of deubiquitinating enzymes (DUBs) that can deubiquitinate ubiquitinated proteins widely in cells.22,23) Human genes encode a total of about 103 recognized DUBs, divided into six different types, which are involved in various stages of ubiquitination modification regulation.24)
USP9X (ubiquitin-specific peptidase 9 X-linked) is an important member of the USP enzyme and belongs to the DUBs family. It can remove ubiquitin molecules from large proteins, thereby inhibiting the degradation of target proteins and performing important protein-regulatory functions.25) Recent studies have shown that USP9X could affect different biological processes by stabilizing different substrates, such as protein transport, apoptosis, polarization, autophagy, cell growth and migration, immune response, and self-renewal and differentiation of stem cells.26) USP9X also participated in a variety of signal pathways. USP9X was an important part of transforming growth factor β (TGF-β) signaling pathway; it could participate in the regulation of mitogen-activated protein kinase (MAPK) signaling pathway; it mediated the activation of apoptotic signal regulating kinase 1 (ASK1) signaling pathway by inhibiting the degradation of ASK 1.27) However, USP9X is involved in extracellular signal-regulated kinase 1/2 (ERK1/2) and phosphatidylinositol-3-kinases/protein-serine-threonine Kinase (PI3K/Akt) Pathway is rarely studied.
USP9X has been reported to be abnormally expressed in various diseases, including many cancers.25,28) However, the mechanism of action of USP9X in the development and progression of osteosarcoma remains to be studied. In this study, we report that down-regulation of USP9X can inhibit the proliferation, migration and invasion of osteosarcoma.
Tumor and adjacent tissue specimens from 67 osteosarcoma patients (mean age, 22.25 ± 4.05 years; 40 males and 27 females) were obtained between June 2015 and June 2018 at the Henan Provincial People’s Hospital. Paracancer tissues were collected at the same time as osteosarcoma tissue samples after surgical resection, and were collected at least 3 cm away from the edge of tumor tissues. The clinical data of the patients are listed in Table 1. After removing the tissue specimens, immediately freeze them with liquid nitrogen and store them at −80 °C for further experiments. The Ethics Committee of Henan Provincial People’s Hospital approved the research protocol. Each sample was collected after receiving written informed consent before being anonymously analyzed. This program was carried out in accordance with the “Declaration of Helsinki.” Parents or legal guardians of patients under the age of 18 provided written informed consent.
| Clinical characteristics | Cases (n = 67) | USP9X expression | p-Value | |
|---|---|---|---|---|
| High (n = 34) | Low (n = 33) | |||
| Sex | 0.882 | |||
| Male | 40 | 20 | 20 | |
| Female | 27 | 14 | 13 | |
| Age (years) | 0.724 | |||
| <18 | 29 | 14 | 15 | |
| ≥18 | 38 | 20 | 18 | |
| Tumor site | 0.657 | |||
| Distal femur | 22 | 12 | 10 | |
| Proximal tibia | 28 | 15 | 13 | |
| Others | 17 | 7 | 10 | |
| Tumor size (cm) | 0.274 | |||
| <10 | 39 | 22 | 17 | |
| ≥10 | 28 | 12 | 16 | |
| Enneking staging | 0.005* | |||
| I-II | 37 | 13 | 21 | |
| III | 30 | 24 | 9 | |
| Distant metastasis | 0.016* | |||
| Absent | 41 | 16 | 25 | |
| Present | 26 | 18 | 8 | |
Notes: χ2 test, * p < 0.05.
Each human osteosarcoma tumor specimen was stained with H&E. The slices were placed in a 45 °C-drying oven for 6–8 h. Remove slices after cooling to room temperature, then dewaxing, a series of degreasing and dehydration processes from xylene to ethanol were processed. After completion, hematoxylin was stained for 10 min → 1% hydrochloric acid alcohol differentiation for 30 s → 1% dilute ammonia anti-blue 20 s (Zsbio, China) → eosin staining for 10 min (Zsbio) → dehydrate and seal. The dehydration process is the reverse process of dewaxing. The baking and dewaxing process is the same as that of H&E staining. The subsequent process was antigenic repair → blocking endogenous peroxidase → add the blocking fluid → drops of primary antibody were added and incubated overnight at 37 °C (anti-USP9X, CST, #14898, 1 : 1000) → biotin-labeled rabbit immunoglobulin G (IgG) polymer was added → drops of horseradish enzyme-labeled streptomycin working fluid were added → dimethylaminoazobenzene (DAB) (Zsbio) chromogenic → re-staining with hematoxylin → dehydrate and seal (same with H&E staining). All scans were performed using Aperio AT2 (Leica, U.S.A.). The cutoff value was sorted from high to low, the top 50% (34 cases) of USP9X showed high expression, and the remaining 33 cases showed low expression.
Cell Lines and Cell CultureMouse osteosarcoma cells lines K7M2 and human osteosarcoma cells MG-63 were purchased from Chinese Academy of Sciences, Shanghai Cell Bank. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (BI, Israel) with 10% fetal bovine serum (FBS, BI) and incubated in a CO2 incubator at 37 °C with 95% humidity.
Thiazolium Blue (MTT) AssayThe density of cells was 3 × 103/well, seeded on 96-well plates, discard the original medium when the cell adheres to the wall, and added fresh medium contained different concentrations of USP9X inhibitor. Then discarded original medium and add 150 µL dimethyl sulfoxide (DMSO), shaken for 10 min to dissolve the precipitate completely. Measured the absorbance with a microplate reader at 490 nm wavelength, and analyzed with GraphPad 5.0 software.
Sulforhodamine B (SRB) StainSRB satin was used to detect cell viability in different groups. 3 × 103 cells were plated into 96-well plate, 24 h later, replase with fresh medium with or without tested inhibitor and fixed by 10% trichloroacetic acid (TCA) at different time points. Stained with 0.4% SRB and solved at 10 mmol/L tris solution. Optical density for SRB assay was detected at 562 nm by the microplate analyzer.
Wound Healing AssayThe wound healing experiment was used to monitor cell migration ability on a 6-well plate. When the cell fusion rate reached 90%, a straight line at the bottom of the cell using a 200 µL spear tip was drawn. Photographs were taken under an inverted microscope at 0 and 24 h, respectively. The migration rate was analyzed by Image J softwore.
Cell TransfectionThe sequence of USP9X and negative control small interfering RNA (siRNA) were shown in below: USP9X-1 (sense: 5′-AGA AAU CGC UGG UAU AAA UUU-3′; antisense: 5′-AUU UAU ACC AGC GAU UUC UUU-3′), USP9X-2 (sense: 5′-GCA GUG AGU GGC UGG AAG UTT-3′; antisense: 5′-ACU UCC AGC CAC UCA CUG CTT-3′), USP9X-3 (sense: 5′-GGA CUU CUU UGA AAG UAA UTT-3′; antisense: 5′-AUU ACU UUC AAA GAA GUC CTT-3′), negative control (sense: 5′-UUC UCC GAA CGU GUC ACG UTT-3′; antisense: 5′-ACG UGA CAC GUU CGG AGA ATT-3′). According to the manufacturer’s protocol, the cells were plated and grown to 60–80%, and then transiently transfected with H4000.
Transwell Migration AssayThe upper chamber of 1 × 104 cells was incubated in 300 µL medium without fetal bovine serum, and the lower chamber was incubated in 600 µL containing 20% FBS. All cells were hatched in a 37 °C temperature for 48 h. Scraped the uninfected upper chamber cells with a cotton swab, washed twice with phosphate buffered saline (PBS), fix with methanol for 20 min, and then air dry at room temperature after washing.
RNA Isolation and RT-PCR AssayAfter the total RNA was extracted, primescript®RTreagentKit was reverse transcribed into cDNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) controled the level of mRNA expression. The primer sequence was as follows: USP9X (Forward: 5′-CAA TGG ATA GAT CGC TTT ATA-3′; Reverse: 5′-CTT CTT GCC ATG GCC TTA AAT-3′), GAPDH (Forward: 5′-CGG AGT CAA CGG ATT TGG TCG TAT-3′; Reverse: 5′-AGC CTT CTC CAT GGT GGT GAA GAC-3′).
Western BlotThe extracted cells were lysed on ice in radio immunoprecipitation assay (RIPA) buffer containing 1% protein kinase A (PKA), PKB, phenylmethylsulfonyl fluoride (PMSF) and holoenzyme inhibitor for 30 min. Using the bicinchoninic acid (BCA) protein quantification kit to detect the protein concentration. Separate the same amount of protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transfer the corresponding protein to the nitrocellulose filter (NC) membrane. After blocking for 2 h at room temperature, incubated overnight with the primary antibody at 4 °C, and then incubated with the secondary antibody for 2 h at room temperature. Observe the WB band with the enhanced chemiluminescence kit and project it on the X-ray film.
Tumor Xenograft AssayThe Animal Experiment Committee of Henan Provincial People’s Hospital approved this animal experiment. And all the experiments were followed by the NIH laboratory animal guidelines. Four–6 weeks female Balb/c nude mice (Hunan SJA Laboratory Animal Co., Ltd., China) were purchased. Osteosarcoma cells (1 × 107) were inoculated subcutaneously into the right axilla of mice, and the mice were raised until the tumor volume reached 100 mm3. They were divided into 5 mg/kg EOAI3402143 group and control group randomly. During the experiment, the body weight and tumor volume were measured every two days, and the mice were sacrificed 21 d later.
Statistical AnalysisThe measurement data were expressed as mean ± standard deviation (S.D.). All statistical analyses were performed using Graph Pad and SPSS software. The Students’ test method was used to analyze the statistical significance of the differences between the two groups, and the variance analysis method was used to analyze the differences between three groups and above. Log rank test was used to compare the overall survival between 2 groups in Kaplan–Meier analysis. The difference of p < 0.05 was considered significant.
Sixty-seven cases of osteosarcoma and paracancerous tissue samples were collected from our hospital. H&E staining and immunohistochemistry showed that USP9X was mainly expressed in the cytoplasm, and the expression intensity of USP9X in the normal group was lower than that in the tumor group (Fig. 1A). In the Gene Expression Analysis Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn), we evaluated their expression in cancer tissues and para-position osteosarcoma cancer tissues, and the results showed that USP9X in paracancer tissue was also lower than that of cancer tissue (Fig. 1B). The expression of USP9X in osteosarcoma tissue showed an up-regulation trend (Fig. 1C). Furthermore, we found that USP9X expression was correlated with enneking stage (p = 0.005) and metastasis (p = 0.016) (Table 1). the expression of USP9X was not different from the patient’s gender (p = 0.882), age (p = 0.724), tumor location (p = 0.657), tumor size (p = 0.274) in Table 1. In Fig. 1D, Kaplan–Meier’s study shows that the up-regulation of USP9X expression is associated with poor patient survival (p = 0.002). The median overall survival of patients with up-regulation group was significantly lower than that of down-regulation group (31.2 ± 20.5-month vs. 39.4 ± 25.8-month, p = 0.026). Taken together, these results presented that USP9X was involved in the occurrence and development of osteosarcoma.

(A) Sixty-seven patients with osteosarcoma were divided into paraffin-matched tissue sections, which were examined by USP9X IHC (Immunohistochemistry) and H&E (hematoxylin-eosin) staining. (B) GEPIA data showed that USP9X expression was higher in sarcomas than in normal tissues. (C) IHC score showed that USP9X expression was significantly higher in osteosarcoma than in its matched normal tissues. (D) The cumulative survival of patients with high expression of USP9X was significantly lower than that of patients with low expression. TPM (Transcripts per million). Scale bars (the large one is 3 mm and the small one is 200 µm). ** p < 0.01.
We tested the expression of USP9X in mouse osteosarcoma cell K7M2 and human osteosarcoma cell lines Saos2, MG-63 and U2OS (Fig. 2A). RT-PCR and Western blotting results showed that USP9X was highly expressed in K7M2, Saos2 and MG-63 cell lines than in the U2OS cell line (Figs. 2B, C). Therefore, mouse K7M2 and a human MG-63 cell line were selected in our subsequent studies, with high expression of USP9X. In order to explore whether the inactivation of USP9X was sufficient to inhibit the growth of osteosarcoma cells, we selected USP9X inhibitor (EOAI3402143, Fig. 2D) to evaluate the antiproliferative activity in osteosarcoma cells. We found that with the increase of USP9X inhibitor concentration, the growth of K7M2 and MG-63 cells were significantly inhibited (Fig. 2E). We further tested the IC50 of the USP9X inhibitor for both, and the results were 1.54 µmol/L (K7M2, Fig. 2F) and 1.50 µmol/L (MG-63, Fig. 2G), respectively.
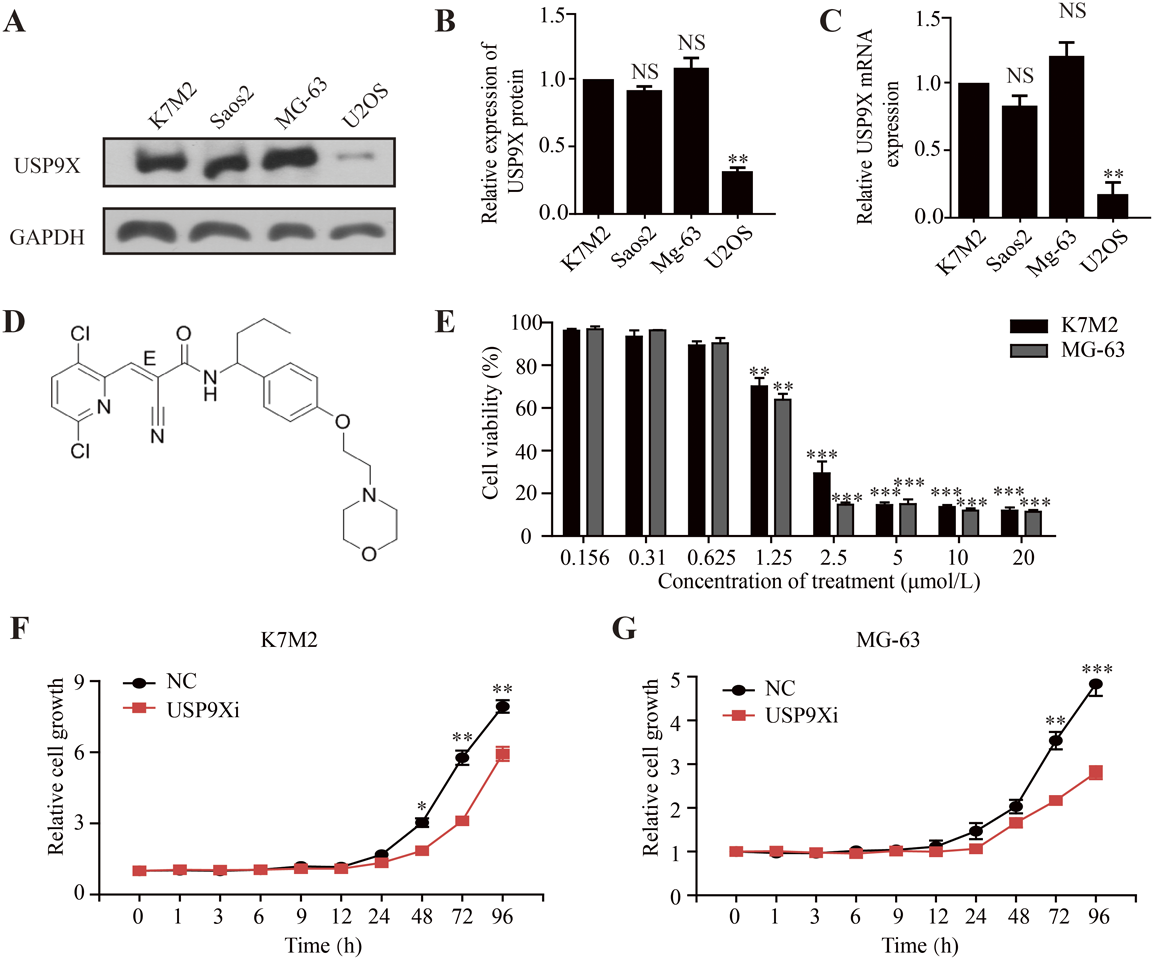
(A) Expression levels of USP9X in four osteosarcoma cell lines as indicated. (B) The protein expression level of USP9X was detected by Western blot assay in osteosarcoma cell lines. (C) The mRNA expression level of USP9X gene was detected by RT-PCR assay in osteosarcoma cell lines. (D) Chemical structure of USP9X inhibitor (EOAI3402143). (E) K7M2 and MG-63 cells were treated with USP9X inhibitor of gradient concentration for 72 h, respectively, and the survival rate of the corresponding cells was detected by MTT (Thiazolium blue) assay. Osteosarcoma cells K7M2 (F) and MG-63 (G) were treated with USP9X inhibitor (500 nmol/L) for different time, and the corresponding cell proliferation was detected by SRB (Sulforhodamine B) staining. USP9Xi, USP9X inhibitor. NS, no statistical significance. ** p < 0.01, *** p < 0.001.
To evaluate the in vivo anti-cancer activity of USP9X inhibitor, we subcutaneously transplanted K7M2 and MG-63 cells into nude mice. Then the USP9X inhibitor was injected into mice and the inhibitor significantly suppressed K7M2 and MG-63 cells’ tumor growth (Figs. 3A, B). Similarly, as shown in Figs. 3C and D, tumor volume in K7M2 and MG-63 mice was limited compared to the negative control group. Considering the above results, we believe that the USP9X inhibitor inhibited the growth of osteosarcoma cells in vitro and in vivo.
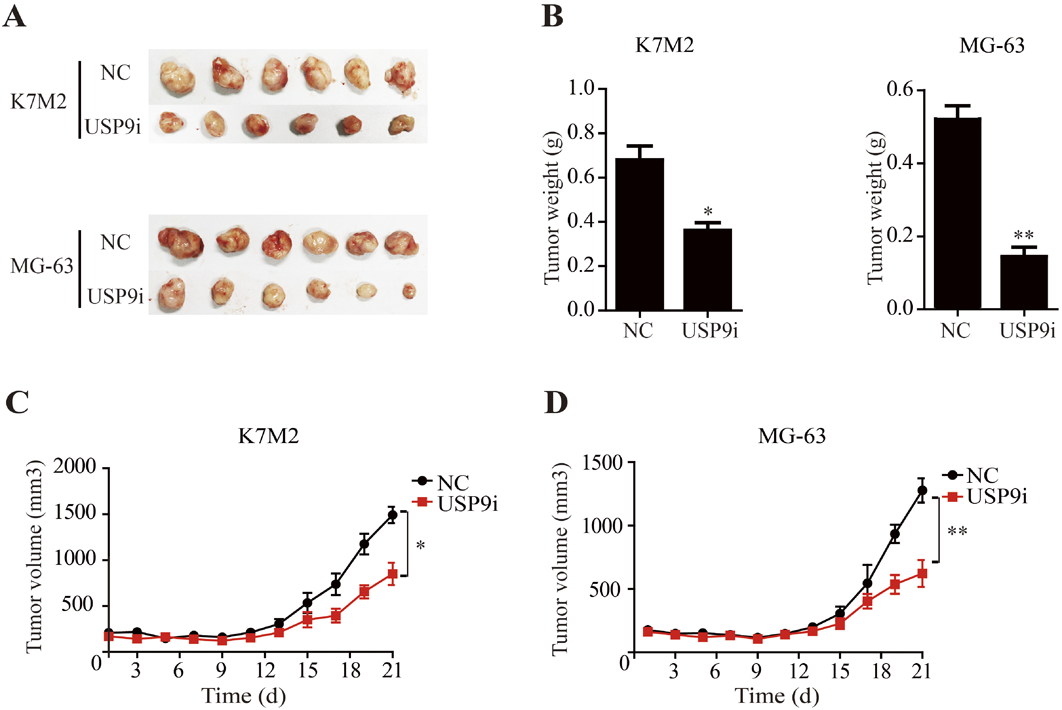
(A) Representative photographs of K7M2 and MG-63 tumors in nude mice. (B) Bar graph represents the results of the average tumor weight (presented as the mean ± standard deviation. Curve of the tumor volume (mm3) of mice in K7M2 (C) and MG-63 (D). * p < 0.05, ** p < 0.01.
Western blotting and RT-PCR were used to detect the transfection efficiency of siRNA-USP9X in osteosarcoma cells. The results showed that under the action of si-USP9X#1, si-USP9X#2 or si-USP9X#3, SRPX2 protein and mRNA levels in K7M2 and MG-63 cell lines were significantly reduced. But it’s more obvious in #1 and #3, where the transfection efficiency of was <30% (Figs. 4A, B, 3C, D). We then observed the effect of down-regulation of USP9X on the migration and invasion of osteosarcoma cells. First, we observed the effect of down-regulation of USP9X on migration of K7M2 and MG-63 cells by cell scratch assay. We found that down-regulation of USP9X significantly inhibited migration of K7M2 and MG-63 cells (Figs. 5A, B). Down-regulation of USP9X could affect the proliferation of osteosarcoma cells. Therefore, transwell assay was conducted to explore further effect of USP9X on the invasion ability of osteosarcoma cells. These showed that down-regulation of USP9X significantly inhibited the invasion of K7M2 and MG-63 cells (Figs. 6A, B). Thus, we believe that down-regulation of USP9X suppresses osteosarcoma cells proliferation migration and invasion in vitro.
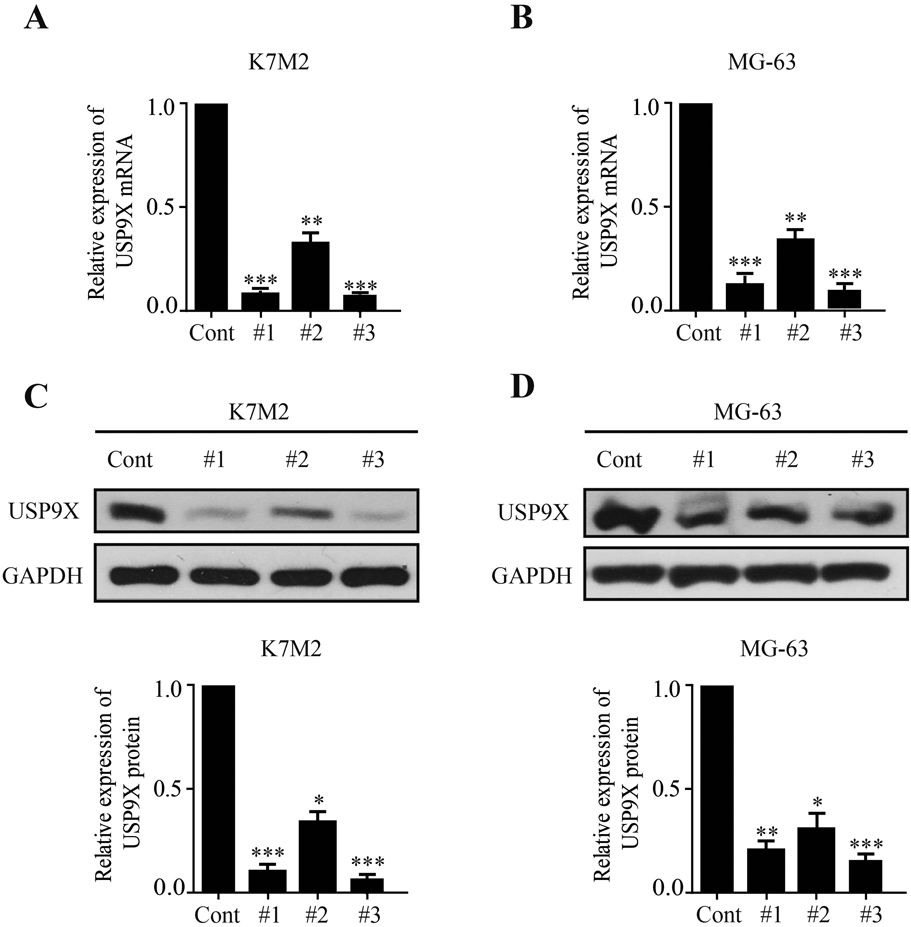
USP9X mRNA level of si-USP9X#1, si-USP9X#2 and si-USP9X#3 groups in K7M2 (A) and MG-63 (B) cells were detected by RT-PCR assay. USP9X protein level of si-USP9X#1, si-USP9X#2 and si-USP9X#3 in K7M2 (C) and MG-63 (D) were detected by Western blot assay. * p < 0.05, ** p < 0.01, *** p < 0.001.
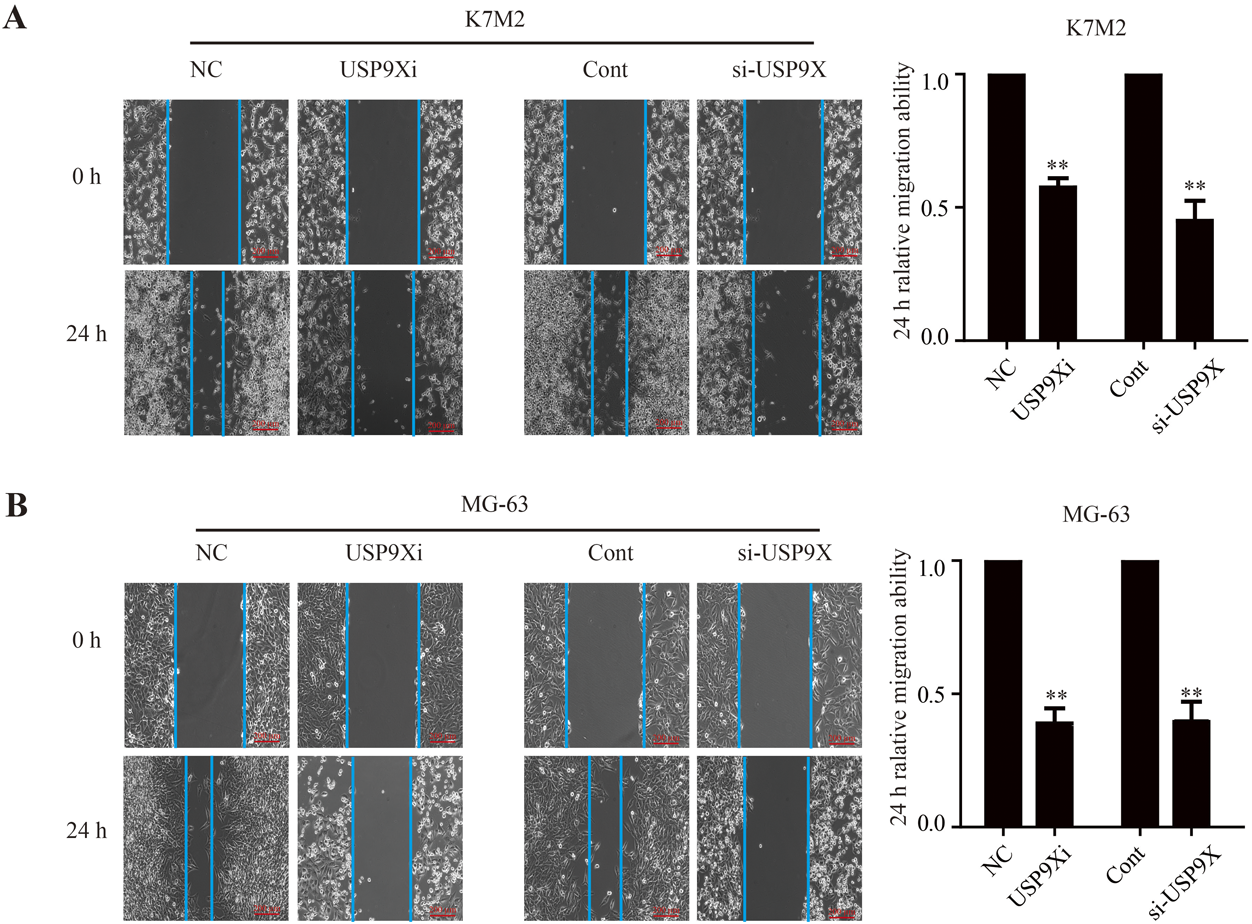
The effect of down-regulation of USP9X on the migration of K7M2 (A) and MG-63 (B) cells was observed by cell scratch assay (scale bars = 200 µm). USP9Xi, USP9X inhibitor. ** p < 0.01.
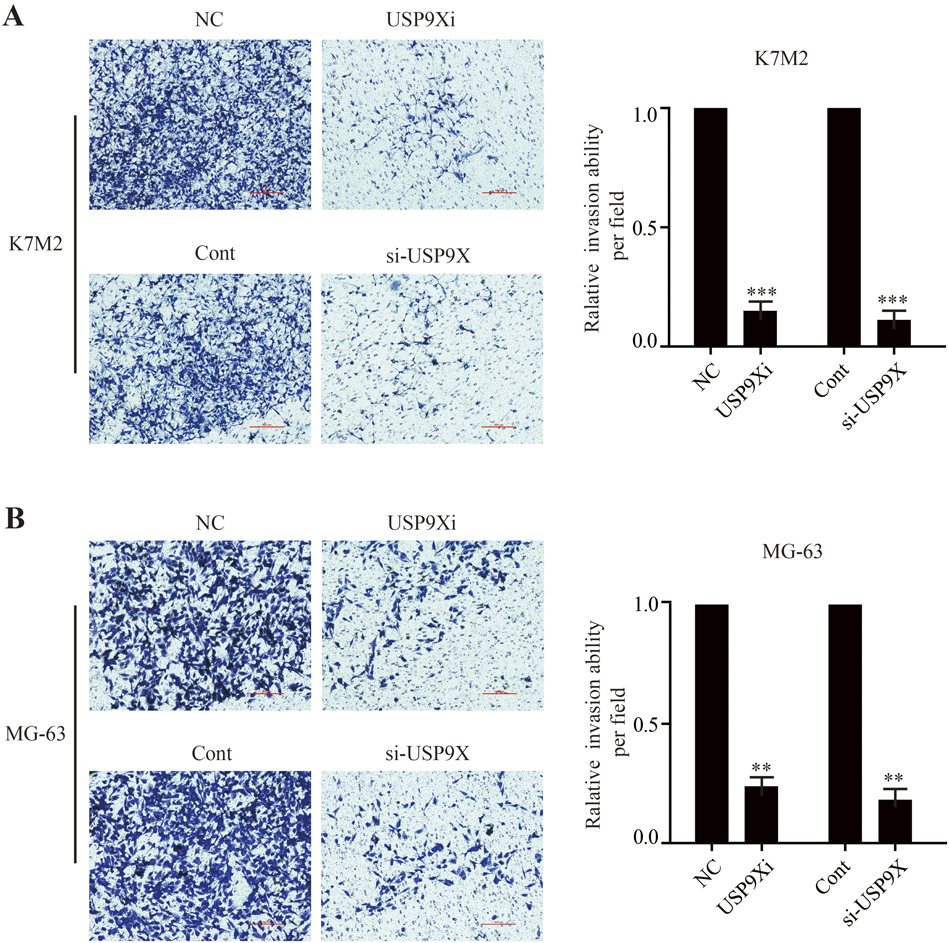
Transwell assay was conducted to observe the effect of down-regulation of USP9X on the invasion ability of K7M2 (A) and MG-63 (B) cells (scale bars = 1000 µm). USP9Xi, USP9X inhibitor. ** p < 0.01, *** p < 0.001.
We further tested the related regulatory pathways of osteosarcoma cell proliferation, migration and invasion, and clarified the molecular mechanism of USP9X regulating the proliferation, migration and invasion of osteosarcoma cells. In Fig. 7A, down-regulation of USP9X significantly decreased the expression of N-cadherin and vimentin and increased the expression of E-cadherin in K7M2 and MG-63 cells, which played an important role in cell migration and invasion.

(A) Protein expression of migration and invasion-related molecules according to Western blot analysis. (B) Western blotting analysis of ERK1/2 and PI3K/Akt signaling-related proteins in K7M2 and MG-63 cell lines. NS, no statistical significance. #1, si-USP9X#1. #3, si-USP9X#3. * p < 0.05, ** p < 0.01, *** p < 0.001.
Erk1/2 had been considered as a signaling pathway related to cancer proliferation and metastasis in many studies.29,30) The PI3K/Akt pathway was also the main signaling pathway related to cancer progression and invasion.31,32) Therefore, we next explored the expression of molecules related to ERK1/2 and PI3K/Akt signaling. In Fig. 7B, p-ERK1/2 and p-Akt in the si-USP9X group were lower than those in the control group of K7M2 and MG-63 osteosarcoma cells. The expression levels of ERK1/2 and Akt were not changed. These results indicated that USP9X was involved in the ERK1/2 and PI3K/Akt pathways of osteosarcoma cells.
Conflicting reports on the role of USP9X as a tumor suppressor or oncogene suggest that USP9X plays a complex role in carcinogenesis. Although several reports had identified USP9X as an oncogene in prostate,33) lymphoma34) and colorectal cancer,35) two studies had shown that interfering with USP9X expression promotes faster onset of pancreatic ductal adenocarcinoma (PDAC) in genotypic mouse models.36,37) And studies had demonstrated that USP9X presents as a proto-oncogene in the context of established PDAC and xenograft models, which is consistent with findings in other tumors.34) TGF-β, for example, appears as a tumor suppressor in the early stages of some tumors, but as a proto-oncogene in the late stages.38) USP9X may behave similarly to TGF-β in some cancers, but whether this paradigm holds true for tumorigenesis of USP9X requires further characterization. The abnormal expression of USP9X played an important role in the formation and development of tumors, and its expression level was closely related to the clinical and pathological characteristics of tumors, as well as the prognosis of tumor patients. Clinically, the detection of USP9X protein expression level might be used to evaluate the prognosis of tumor patients or guide further treatment. However, it should also be clearly recognized that USP9X could promote both apoptosis and cell survival in vitro, which was closely related to the interactions or dominant substrates in different types and states of cells or tumors.39)
There are about 600 genes encoding numerous E3 ligases in human cells, compared with only 95 putative DUBs.40) Thus, each DUBs may remove ubiquitin links on multiple proteins. For example, the deubiquitin enzyme USP7 has been reported to remove polyubiquitin chains on tumor suppressor genes p53, PTEN and FOXO (or proto-oncogenes MDM2), PHF8 and UHRF1, and thus plays an important role in tumor genesis and development.41) USP9X can deubiquitinate various substrate proteins in different signaling pathways and even in fusion signaling pathways and ultimately affect the development of tumors. It’s worth noting that knockdown or inhibition of USP9X can reverse cisplatin resistance in other carcinoma cells.42,43) Furthermore, cisplatin is a key-chemotherapeutic agent in osteosarcoma patients. Thus, it is very necessary to investigate whether USP9X participates in the progress of osteosarcoma.In this paper, first of all, immunohistochemical staining indicated that USP9X was highly expressed in osteosarcoma tissues and was positively correlated with the grade of osteosarcoma malignancy and metastasis. Furthermore, high expression of USP9X in osteosarcoma is associated with poor prognosis. Then, the GEPIA database and Western analysis revealed that USP9X mRNA expression levels and protein levels were upregulated in osteosarcoma. Simultaneously, we performed tumor formation experiments in nude mice, and found that the tumor formation rate of osteosarcoma was significantly inhibited in mice injected with USP9X inhibitor. To identify the pathways involved in the down-regulation of USP9X to inhibit proliferation, migration, and invasion of osteosarcoma cells, we further examined the pathway proteins associated with osteosarcoma.Moreover, p-ERK1/2 and p-Akt were up-regulated.
In summary, USP9X was overexpressed in osteosarcoma tissues and cells. The proliferation, migration and invasion of osteosarcoma cells were inhibited by the down-regulation of USP9X, which was related to the ERK1/2 and PI3K/Akt signaling pathways. In future studies, we will further explore the mechanism of USP9X influencing the development of osteosarcoma, so as to lay a solid foundation for translational therapy of osteosarcoma.
This work was supported by National Natural Science Foundation of China (No. 82002840) and Key Scientific and Technological Projects in Henan Province (LHGJ20210024).
The authors declare no conflict of interest.