2023 年 46 巻 1 号 p. 74-85
2023 年 46 巻 1 号 p. 74-85
Breast cancer (BC) is the most generally diagnosed cancer and the driving cause of cancer-related death. Transmembrane (TMEM) proteins have been reported to serve as prognostic indicators in a variety of cancers, and it can offer therapeutic targets for carcinoma. However, the function of TMEM in BC remains unclear. In this study, TMEM9A, a member of TMEM family, was screened as the candidate gene after analyzing the profiles of GSE42568 and GEPIA-BRCA database via bioinformatic method. The upregulated expression of TMEM9A was confirmed in BC samples compared with the paired normal tissues. Hence, we speculated that TMEM9A might promote BC progression. To test the hypothesis, we performed a series of loss/gain-of-function experiments and found that BC cells with TMEM9A deletion inhibited cell proliferation, migration, and invasion along with induced apoptosis. Conversely, TMEM9A overexpression reversed the trend. Mechanically, TMEM9A knockdown blocked the Wnt/β-catenin signaling pathway as evidenced by the increased adenomatous polyposis coli (APC) expression and decreased β-catenin, cyclin D1, and axis inhibition protein 2 (AXIN2) expression. Furthermore, over-activation of the Wnt/β-catenin pathway by transfecting BC cells with β-Catenin-S33Y (β-Catenin tyrosine for serine at codon 33) plasmids reversed the effects caused by TMEM9A knockdown. In conclusion, TMEM9A may play a tumor-promoting role in BC progression via activating the Wnt/β-catenin signaling pathway. Therefore, TMEM9A may be an effective therapeutic option for BC.
Breast cancer (BC) is the most common cancer affecting women and ranks first for incidence in the vast majority of countries.1,2) Current evidence suggests that BC in women aged <45 years is the leading cause of cancer-related deaths.3) According to the report, 20–30% of BC patients may undergo metastasis after tumor resection and adjuvant therapy of the primary tumor.4) Although researchers have made great strides in the treatment and diagnosis of BC,5,6) BC is still a major health issue and currently represents a top biomedical research priority.
Transmembrane proteins (TMEMs) contains an N-terminal signal peptide, a single transmembrane region, three potential N-glycosylation sites, and three conserved cysteine (Cys)-rich domains in the N-terminus.7) TMEM9A, the member of TMEMs family, encodes a 183 amino-acid protein and is highly conserved between species from Caenorhabditis elegans to humans.7) Previous studies indicated that TMEM9A was abnormally expressed in different cancers. For example, TMEM9A was highly expressed in colorectal cancer,8) and it was revealed to be upregulated in regenerating liver and hepatocellular carcinoma cells.9) Furthermore, TMEM9A exerted a tumor-promoting role in human hepatoma cell lines, and TMEM9A knockdown could notably inhibit the proliferation, invasion, migration, and induce apoptosis.10) Another report suggested that TMEM9A could promote intestinal tumorigenesis and hyperactivated the expression of β-catenin.8) Emerging evidence illustrated that TMEM9A was tightly ssociated with cancerous development and exacerbated the malignant behavior of tumors. In this study, bioinformatic data analyzed that TMEM9A expression was upregulated in BC tissues, and we also identified that the transcriptional and protein levels of TMEM9A were prominently increased in BC tissues compared with adjacent normal tissues. However, the function and mechanism of TMEM9A in BC were not well defined.
The Wnt/β-catenin signaling pathway as a conserved signaling pathway is revealed to involve in diverse physiological processes, such as proliferation, differentiation, apoptosis, migration, invasion, and tissue homeostasis.11,12) In the absence of Wnt/β-catenin signal, β-catenin is phosphorylated and hijacked by the destruction complex, which comprises the scaffolding protein Axin, adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK-3β), and casein kinase 1α (CK1α).13–16) Subsequently, the phosphorylated β-catenin is recognized and degraded by a ubiquitin ligase-proteasome system.17) Conversely, the Wnt/β-catenin signaling pathway is activated by truncated APC protein, which negates the complex-mediated destruction of β-catenin ubiquitination.18) Once unphosphorylated β-catenin enters the nucleus, it initiates the transcription of downstream genes. Moreover, the Wnt/β-catenin signaling pathway is closely related to the tumorous initiation and progression, which includes cell proliferation and differentiation.19) Thus this pathway exerts important roles in carcinogenesis and therapy response. Khramtsov et al. indicate that wnt/β-catenin pathway activation is enriched in BC and this pathway predicts a lower overall survival rate.20) Therefore, the Wnt/β-catenin pathway is deemed an attractive target for this aggressive BC subtype. In addition, overexpression of TMEM9A enhances the protein expression of the canonical Wnt/β-catenin.21) TMEM9A can activate the Wnt/β-catenin pathway to facilitate liver regeneration and tumorigenesis.9) These findings imply that TMEM9A is closely associated with Wnt/β-catenin signaling, which triggers us to explore whether TMEM9A is involved in the progression of BC by controlling the Wnt/β-catenin pathway.
The BC dataset (GSE42568, including 104 breast cancer and 17 normal breast biopsies)22) was downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geoprofiles).23) The Gene Expression Profiling Interactive Analysis (GEPIA) database included BC samples and normal samples was downloaded from GEPIA (http://gepia.cancer-pku.cn).24) In GSE42568, GEO2R tool (http://www.ncbi.nlm.nih.gov/geo/geo2r)25) was used to screen differentially expressed genes (DEGs). For volcano analysis, the standard for screening DEGs was |log2 fold change (FC)| >1.2 and adjusted p < 0.001. Overlapped upregulated genes were screened with log2FC > 1.2 and adjusted p < 0.001 by integrated data, and downregulated genes were screened by the standard of log2FC <−1.2 and adjusted p < 0.001. Overlapped genes were respectively shown in corresponding Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/). The cluster Profiler R package26) was used to perform the histograms of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) in the overlapped genes. The DEGs in TMEM family were screened using the data of GSE42568 and GEPIA. The TMEM9A expression in human BC tissues was analyzed by the TNMplot bioinformatic tool (https://tnmplot.com/analysis/).27) TMEM9A expression in various cancer types was performed by Gene Set Cancer Analysis (http://bioinfo.life.hust.edu.cn/GSCA/#/).28)
BC Tissue SpecimensThirty-seven pairs of para-carcinoma tissues and BC tissues were collected from BC patients as the clinical specimens. Each participant provided informed consent in the study. This study was approved by the Research Ethics Committee of the Fourth Hospital of Hebei Medical University (No: 2020KY113), and this study was carried out following the ethical principles of the latest version of the Declaration of Helsinki.
Cell Culture and TransfectionHS-578T, MDA-MB-231, MDA-MB-468, MCF-7, and MCF-10A cells were purchased from iCell Bioscience (China). T-47D cells were ordered from Procell (China). HS-578T cells and T-47D cells were cultured in Dulbecco's modified Eagle medium and Roswell Park Memorial Institute-1640 medium respectively. Leibovita’s L-15 medium was used to culture MDA-MB-231 and MDA-MB-468 cells. MCF-7 cells were cultured in the minimal essential medium. All of the media were supplemented with 10% fetal bovine serum (FBS) (Tianhang Biotechnology, China) and the above cells were cultured in an incubator at 37 °C and 5% CO2.
T-47D cells were transfected with TMEM9A-specific small interfering RNAs (siRNAs) or the negative control (NC) using Lipofectamine 3000 (Invitrogen, U.S.A.) according to the manufacturer’s protocol. siRNA sequences used in this study were listed in Table 1. The coding sequence (CDS) of TMEM9A was constructed to the plasmid pDONR223 (Youbio Biotechnology, China) to achieve TMEM9A overexpression (TMEM9A-OE). The constructed vector of TMEM9A-OE or empty vector (EV) were transfected into MDA-MB-231 cells using Lipofectamine 3000. For experiments of co-transfection, the reporter plasmids β-Catenin-S33Y (β-Catenin tyrosine for serine at codon 33) or EV and TMEM9A siRNA-4 were co-transfected into T-47D cells, which were named siTMEM9A+β-Catenin-S33Y and siTMEM9A+EV respectively. The transfected cells were used in the follow-up experiments after 48 h incubation.
| Name | Sequence (5′–3′) | |
|---|---|---|
| siTMEM9A-1 | Sense | GCAGCACCACCACCAUCAATT |
| Antisense | UUGAUGGUGGUGGUGCUGCTT | |
| siTMEM9A-2 | Sense | GGUGGAAGCUGCAGGUGCATT |
| Antisense | UGCACCUGCAGCUUCCACCTT | |
| siTMEM9A-3 | Sense | AGGUCAUCAUUGUCAUCUATT |
| Antisense | UAGAUGACAAUGAUGACCUTT | |
| siTMEM9A-4 | Sense | GGCACAAGAUGCUCAGCUATT |
| Antisense | UAGCUGAGCAUCUUGUGCCTT | |
| siTMEM9A-5 | Sense | GGAUGCAUAUACUGAGCAATT |
| Antisense | UUGCUCAGUAUAUGCAUCCTT | |
| siNC | Sense | ACGUGACACGUUCGGAGAATT |
| Antisense | UUCUCCGAACGUGUCACGUTT |
Total RNA was isolated by TRIpure lysis and reverse-transcribed using BeyoRT II M-MLV reverse transcriptase (Beyotime, China) to synthesize cDNA. For RT-PCR, PCR conditions were 94 °C for 5 min and 35 cycles at 94 °C for 20 s, 55 °C for 20 s and 72 °C for 30 s, followed by a final extension step at 72 °C for 10 min. PCR products were then electrophoresed through a 1.5% agarose gel. The mRNA level of TMEM9A was performed by the gel imaging system. For quantitative RT-PCR, SYBR Green dye (Solarbio, China) was used to measure amplification. The mRNA levels of TMEM9A, cyclin D1, and axis inhibition protein 2 (AXIN2) were analyzed using the 2−ΔΔCT method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as endogenous control. The sequences of primers were listed in Table 2.
| Name | Sequence (5′–3′) | |
|---|---|---|
| TMEM9A | Forward | CTTTGGTGGCTGTGGTC |
| Reverse | CTGGGATACATTCTGGTTG | |
| cyclin D1 | Forward | GCGAGGAACAGAAGTGCG |
| Reverse | TGGAGTTGTCGGTGTAGATGC | |
| AXIN2 | Forward | ACCGTGGTTGGCTTGTC |
| Reverse | TCTACACTGCTGTCCGTCAT | |
| GAPDH | Forward | GACCTGACCTGCCGTCTAG |
| Reverse | AGGAGTGGGTGTCGCTGT |
The whole-cell lysates were collected in radio immunoprecipitation assay (RIPA) lysis buffer (Solarbio, China). The protein concentration was determined using a BCA Protein Assay Kit (Solarbio, China). Equal amounts of protein were separated by electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, U.S.A.). After blocking with 5% non-fat milk, the membranes were incubated with specific primary antibodies against TMEM9A (Proteintech, China, catalog number: 19918-1-AP, 1 : 1000); APC (Abclnoal, China, catalog number: A17912, 1 : 500); β-catenin (Abclnoal, catalog number: A19657, 1 : 1000); active-β-catenin (CST, China, catalog number: #8814, 1 : 500); GAPDH (Proteintech, catalog number: 60004-1-Ig, 1 : 10000) overnight at 4 °C and subsequently incubated with horseradish peroxidase-conjugated secondary antibody immunoglobulin G (IgG)-horseradish peroxidase (HRP) (Solarbio, catalog number: SE134, 1 : 3000) at 37 °C for 1 h. western blots were detected using ECL Detection Reagents (Solarbio).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT)Cells were seeded to a 96-well plate with 7 × 103 cells per well. After 48 h-transfection, the 96-well plates were cultured at 37 °C and 5% CO2 for 0, 24, 48, 72, or 48 h. MTT solution (10 µL, Solarbio) was added to the plate and incubated for 4 h. After removing the supernatant, dissolved in dimethyl sulfoxide (110 µL, Sigma, U.S.A.) was added to dissolve the purple crystals in the dark for 10 min. The optical density was measured on the microplate tester (Biotek, U.S.A.) at 490 nm.
Flow Cytometry AssayFor cell cycle analysis, the transfected cells were fixed with cold 70% ethanol at 4 °C overnight. The cell cycle was detected by a Cell Cycle Analysis Kit (Biosharp, China) according to the manufacturer's protocols. Briefly, the cells were washed with phosphate buffer saline (PBS) and then stained with propidium iodide (PI) at 37 °C for 30 min in the dark. After incubation, the samples were analyzed by a flow cytometer (ACEA Biosciences, U.S.A.).
An apoptosis assay kit (Biosharp) was used to detect cell apoptosis in transfected BC cells. In brief, the cells were washed with PBS and resuspended with 1× Binding buffer. Annexin V-fluorescein isothiocyanate (FITC) (5 µL) was used to stain for 10 min in the dark. After that, PI staining (10 µL) and PBS (400 µL) were added to the cells. The mixed samples were analyzed using flow cytometry.
Wound Healing AssayAfter 48 h-transfection, MDA-MB-231 or T-47D cells were cultured in serum-free medium (SFM) and treated with 1 µg/mL mitomycin C (MCE, U.S.A.) for 1 h. Subsequently, cells were scratched in a straight line with a 200 µL pipette tip and the cell surface was washed with SFM. Each image for the gaps was photographed at the start (0 h) and endpoint (12 h) of the experiment using a microscope (200× magnifications).
TranswellFor invasion assay, transwell was additionally pre-coated with matrigel (Corning, U.S.A.) diluted in a ratio of 1 : 3 with a SFM on ice. The gel was solidified in a 37 °C incubator for 2 h. The transfected cells for the invasion assay were seeded in a SFM in the upper chamber. The lower chamber was supplemented with 10% FBS. After incubation at 37 °C for 24 h, the invasive cells were fixed with paraformaldehyde (Aladdin, China) and stained with 0.1% crystal violet (Amresco, U.S.A.). Finally, cells were photographed and counted using an inverted microscope at 200× magnification (Olympus, Japan).
Statistical AnalysisThe software GraphPad Prism 8.0 was used for statistical analysis. Statistical significance was performed using the one-way ANOVA and the two-tailed Student's t-test. The results were considered significant at p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). All data were presented as the mean ± standard deviation (S.D.).
The publicly available expression dataset GSE42568 contained the 104 BC and 17 normal breast biopsies were used for bioinformatic analysis. As shown in Fig. 1A, the volcano plot indicated the DEGs in GSE42568 and GEPIA-breast invasive carcinoma (BRCA). The inner track (GSE42568) and outer track (GEPIA) showed the location of the downregulated (green) and upregulated (red) genes on the human chromosome (Fig. 1B). The overlapping upregulated 376 genes and downregulated 587 genes were integrated in GSE42568 and GEPIA, as shown in Venn diagram (Figs. 1C, D). In 376 upregulated genes, GO enrichment analysis were performed, and the terms of top 30 were selected to generate histogram, as shown in Fig. 2A. The KEGG analysis indicated that the upregulated genes were mainly enriched in cell cycle, DNA replication, and bladder cancer (Fig. 2B). The results of GO analysis and KEGG analysis of downregulated genes were shown in Figs. 2C, D. The pathways of KEGG were mainly enriched in peroxisome proliferator-activated receptor (PPAR) signaling pathway, regulation of lipolysis in adipocytes, and pyruvate metabolism.

(A&B) Volcano and circos plots showed the DEGs in GSE42568 and GEPIA-BRCA. (C&D) The upregulated and downregulated genes after integrating GSE42568 and GEPIA. GEPIA-BRCA: Gene Expression Profiling Interactive Analysis-breast invasive carcinoma.

(A&B) The histogram showed GO enrichment and KEGG enrichment in overlapping upregulated genes. (C&D) The histogram showed GO analysis and KEGG analysis in overlapping downregulated genes. GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes. BP: biological process; CC: cellular component; MF: molecular function.
It has been reported that TMEM expression is dysregulated and TMEMs as tumor suppressors or oncogenes exhibit oncogenic properties in different tumors.29) Hereon, the volcano plots showed the gene number of TMEM family in GSE42568 and GEPIA (Supplementary Fig. 1). Then the DEGs in TMEM family were screened by overlapping GSE42568 and GEPIA (Fig. 3A). The heat map showed the gene expression in normal and BC tissues of GSE42568, and the results suggested that the number of up-regulated (red) and down-regulated (blue) genes in overlapping TMEM family was 4 and 6, respectively (Fig. 3B). The circos plot showed the 10 genes in the human chromosome. The expression of TMEM9A was analyzed by the data in GSE42568, and it was shown in the center of the circos plot (Fig. 3C). By analyzing the expression of TMEM9A in BC, we found that TMEM9A was significantly upregulated in tumor tissues compared with tumor-adjacent samples (Fig. 3D) and normal samples (Fig. 3E). Figure 3F showed the dysregulated TMEM9A expression in different cancer types, and we discovered that TMEM9A was upregulated in BRCA samples.

(A) Venn diagram of TMEM family in GSE42568 and GEPIA database. (B) Heatmap showed the differently expressed TMEM membranes between normal tissues and tumor tissues. The blue letter represents the downregulated genes, the red letter represents the upregulated genes. (C) The circular plot of TMEM genes with chromosomes was shown in the outer tracks. The center of the circular plot showed the expression of TMEM9A in GSE42568. (D&E) TMEM9A expression in BC tissues from breast invasive carcinoma compared with the adjacent tumor tissues and normal tissues. (F) TMEM9A expression in different cancer types by GSCA (http://bioinfo.life.hust.edu.cn/GSCA/#/). N: normal; T: tumor.
The increased mRNA level of TMEM9A was validated using qRT-PCR in 37 pairs of para-carcinoma tissues and BC tissues (Fig. 4A). Subsequently, 6 pairs of samples were used for the detection of the transcriptional and protein levels of TMEM9A (Fig. 4B). The results indicated that TMEM9A was profoundly upregulated in BC tissues compared with para-carcinoma tissues. Normal cell line (MCF-10A cells), and BC cell lines (HS-578T, MDA-MB-231, MDA-MB-468, MCF-7, and T-47D cells) were used to identify the mRNA and protein levels of TMEM9A. The results of RT-PCR and Western blot showed that TMEM9A was highly expressed in BC cell lines compared with the MCF-10A line (Figs. 4C, D). To study the biological function of TMEM9A in BC, we transfected TMEM9A-OE and si-TMEM9A to MDA-MB-231 (the lowest TMEM9A expression) and T-47D cells (the highest TMEM9A expression) respectively. The results of qRT-PCR and Western blot verified the transfected efficiency in MDA-MB-231 cells and T-47D cells (Figs. 4E–H). Therein, si-TMEM9A-4 and si-TMEM9A-5 were selected for the followed experiments (Figs. 4F, H).

(A&B) The transcriptional and protein levels of TMEM9A were detected in normal and BC tissues. (C&D) Relative TMEM9A mRNA and protein levels were detected in BC cell lines (HS-578T, MDA-MB-231, MDA-MB-468, MCF-7, and T-47D cells) and normal cell lines (MCF-10A cells) by real-time PCR and Western blot. GADPH was used as the control of sample loading. (E–H) Quantitative RT-PCR was performed to verify the transfected efficiency of TMEM9A in MDA-MB-231 and T-47D cells on mRNA and protein levels. *** represented p < 0.001. TMEM9A, Transmembrane protein 9A. TNMplot, Tumor, Normal and Metastatic plot.
As indicated in Figs. 5A and B, the MTT assay suggested that TMEM9A knockdown notably decreased the cell viability compared with the NC group at 24 h, 48 h, and 96 h, whereas TMEM9A overexpression exerted the opposite effects. The percentage of cells in the G1 phase in the TMEM9A-OE group was markedly decreased, and the cell distribution in the G2 and S phases were increased (Fig. 5C). The flow cytometric analysis revealed that TMEM9A knockdown lead to cell cycle arrest in T-47 cells (Fig. 5D, Supplementary Fig. 2). Also, the apoptotic rate was higher in TMEM9A-silenced cells than in control cells (Fig. 5E).

(A&B) Cell viability of TMEM9A in MDA-MB-231 and T-47D cells with TMEM9A overexpression and deletion was detected at the indicated time by MTT assay. (C&D) Cell cycle distribution was evaluated in TMEM9A-overexpressed and silenced cells by flow cytometry. (E) The apoptosis rate (Q2 + Q4) in TMEM9A-silenced cells was analyzed using flow cytometry. AnnexinV+/PI− (Q4): the early apoptotic cells; AnnexinV+/PI+ (Q2): the late apoptotic cells. **, ***represented p < 0.01, p < 0.001 respectively.
To further probe the impact of TMEM9A on malignant behaviors, we detected the migratory and invasive abilities in BC cells. Wound healing assay showed that TMEM9A overexpression promoted migration, and TMEM9A deficiency prominently inhibited the migratory activity (Figs. 6A, C). The results of the transwell assay suggested that the overexpression of TMEM9A markedly enhanced cell invasion in MDA-MB-231 cells, contrarily, the invasive ability was decreased in the TMEM9A-silenced cells (Figs. 6B, D).

(A–D) The migratory and invasive capacities were evaluated in MDA-MB-231 and T-47D cells through wound healing and transwell assays respectively. The bar graph represented the statistical quantification of migration and invasion in the indicated groups (Scale bar = 100 µm). *, **, *** represented p < 0.05, p < 0.01, p < 0.001, respectively.
Given that the association between the Wnt/β-catenin signaling and carcinoma as well as TMEM9A, we explored the effects of TMEM9A on the Wnt/β-catenin pathway in BC cells. Western blot analysis suggested that TMEM9A overexpression presented the upregulated expression of total and active β-catenin, while the protein level of APC was dropped, and TMEM9A deficiency exerted the opposite effects (Figs. 7A, B). It could be seen from Figs. 7C, E that the mRNA level of cyclin D1 and AXIN2 was increased in TMEM9A-overexpressed MDA-MB-231 cells, whereas TMEM9A deletion decreased the expression of cyclin D1 and AXIN2 in T-47D cells (Figs. 7D, F).

(A&B) Western blot analysis for β-catenin, active β-catenin, and APC in TMEM9A-overexpressed and silenced cells. (C–F) Relative mRNA levels of cyclin D1 and AXIN2 were detected by quantitative RT-PCR. *** represented p < 0.001.
Phosphorylated β-catenin is degraded after it is dissociated from the destruction complex, while un-phosphorylated β-catenin can migrate to the nucleus and accumulate, thereby triggering the Wnt/β-catenin pathway.19,30) T-47D cells with TMEM9A deletion were transfected with the plasmid of β-Catenin-S33Y (non-phosphorylated status) to activate the Wnt/β-catenin pathway. The results of MTT assay suggested that the siTMEM9A + EV group decreased the cell viability, which was reversed by the siTMEM9A + β-Catenin-S33Y group (Fig. 8A). The elevated apoptotic rate in T-47D cells induced by TMEM9A knockdown was suppressed in the presence of β-Catenin-S33Y (Figs. 8B, D). As shown in Figs. 8C and E, the decreased invasive cells after TMEM9A silencing were partially rescued via β-catenin activation.
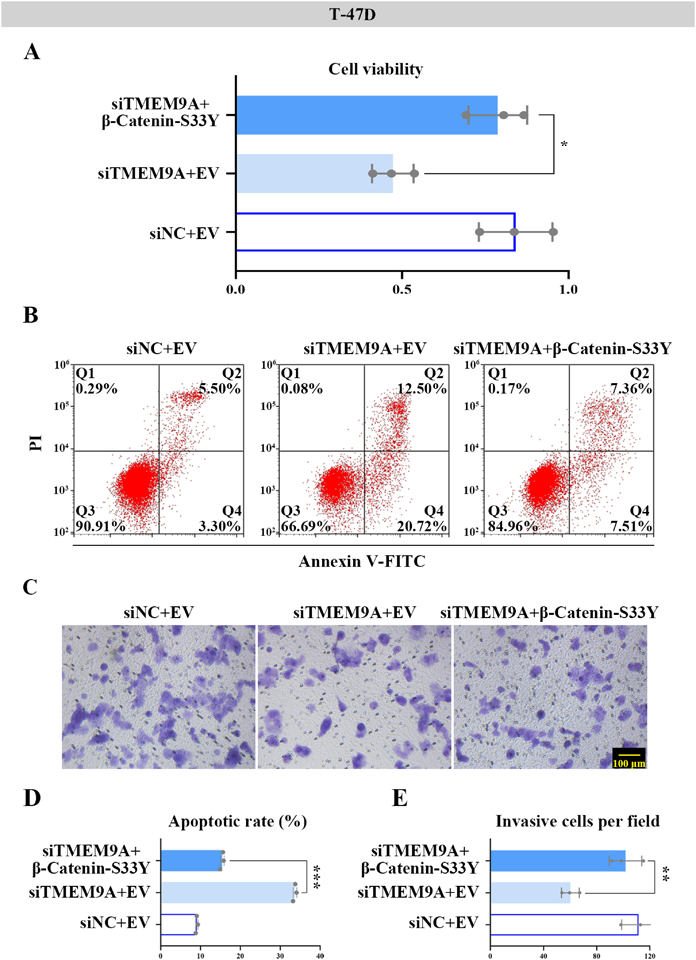
The T-47D cells co-transfected with the vectors of β-Catenin-S33Y and TMEM9A siRNA-4 were used for the following experiments. (A) After 48 h-transfection, cell viability was detected by MTT in T-47D cells. (B) The apoptosis rate (Q2 + Q4) was evaluated by using Annexin V-FITC/PI staining assay. AnnexinV+/PI− (Q4): the early apoptotic cells; AnnexinV+/PI+ (Q2): the late apoptotic cells. (C) The images of invasive cells in indicated groups were captured at 24 h by transwell assay (Scale bar = 100 µm). (D&E) The apoptosis rate and invasive cells were quantified. *, ** represented p < 0.05, p < 0.01, respectively.
TMEMs are reported to be upregulated or downregulated in cancerous tissues compared to adjacent normal tissues.29) TMEMs also participate in drug resistance, and some of TMEMs are used as prognostic biomarkers,29,31) suggesting that TMEM family as a prominent group plays the important role in cancer research. In this study, bioinformatics analysis identified the overlapped DEGs in TMEMs and their expression was shown in the heat map. Hereon, upregulated TMEM family genes were been selected to investigate the tumor-promoting role in BC. Among the four up-regulated genes, TMEM97 is reported to involve in the proliferation and migration of the BC cell lines.32,33) According to the published literatures and the basis of the thesis, we preferentially selected TMEM9A as a candidate gene, and we will further explore the function of the other members of TMEM family in BC in future studies. Studies have demonstrated that the expression of TMEM9A was associated with the function of a series of cancers such as hepatocellular carcinoma and colorectal cancer.8,10) This study summarized the findings and contributions of TMEM9A to BC and demonstrated that TMEM9A was highly expressed in BC tissues and cell lines. Considering that TMEM9A may be a putative oncogene, silencing or expressing its expression would result in therapeutic effects. In this study, the significant expression of TMEM9A confirmed the transfected efficiency, thus, we did not conduct experiments to verify the percentage of cell transfection efficiency. In vitro experiments showed that genetic ablation of TMEM9A inhibited cell phenotypes, whereas TMEM9A overexpression presented the opposite effects. A strong relationship between TMEM9A and malignant behavior has been described in hepatocellular carcinoma.10)
Wnt/β-catenin signaling involves multiple cellular processes, including proliferation, metastasis, immune microenvironment regulation, and phenotype shaping of BC.34–37) Evidence increasingly has demonstrated that the transcription factor β-catenin is a key component of the Wnt/β-catenin signaling pathway and abnormal regulation of β-catenin can lead to the occurrence of early cancer events.19,38,39) Russell et al. have manifested that cyclin D1, the targeting protein of the β-catenin pathway, may promote hepatocyte proliferation.40) Our studies suggested that TMEM9A predominantly affected the mRNA levels of cyclin D1 and AXIN2, the target genes in the downstream Wnt/β-catenin signaling pathway. Additionally, TMEM9A downregulated the protein production of APC and enriched the protein amounts of β-catenin and active β-catenin, uncovering that TMEM9A accelerated the activation of the Wnt/β-catenin pathway. Based on the findings, we characterized the role of the TMEM9A-activated Wnt/β-catenin pathway in BC development. It is well known that β-Catenin-S33Y is insensitive to GSK-3β-mediated phosphorylation and proteasomal degradation.41) Then we found that β-Catenin-S33Y rescued the decreased cell viability and invasion caused by TMEM9A knockdown in BC cells. Loss of TMEM9A increased the apoptotic rate, while the stimulation of β-catenin reversed the status, revealing that TMEM9A could control the Wnt/β-catenin pathway to promote BC development. Researchers have demonstrated that TMEM9A affects liver regeneration by downregulating APC through lysosomal protein degradation, and further hyperactivating the Wnt/β-catenin pathway,9) which is consistent with the present results. Our study also supports evidence from the exploration reported by Jung et al. that TMEM9A facilitates intestinal tumorigenesis by Wnt/β-catenin signaling pathway.8)
Several reviews have comprehensively discussed the important role of the Wnt/β-catenin pathway in tumorous development and therapy.19,42,43) It is well known that inflammation is recognized as a hallmark feature of cancer progression.44) In the last few decades, the impact of inflammation and the immune system on cancerous progression has gained extensive attention. In inflammatory cancers, the Wnt/β-catenin signaling pathway has emerged as a key pathway in malignant biology.45,46) Suryawanshi et al. report the effect on the mutual coexistence and co-dependence of the Wnt/β-catenin signaling pathway along with various inflammatory cascades.47) A novel strategy for cancer prevention and therapy targeting the crosstalk between canonical Wnt/β-catenin and inflammatory signaling cascades has been mentioned.48) The current study indicated that TMEM9A regulated the Wnt/β-catenin pathway to accelerate breast neoplastic progression. Given the connection between inflammation and cancer as well as the Wnt/β-catenin pathway, we have a conjecture that TMEM9A might also affect the occurrence of inflammation. In hepatoma cells, TMEM9A has been demonstrated to mediate interleukin (IL)-6 and IL-1β secretion as well as modulate the Wnt/β-catenin signaling pathway.21) The evidence suggests that the inflammatory components are involved in neoplastic processes, including proliferation, survival, invasion, and migration, making inflammation serves the primary target for cancer prevention and treatment.48) As a novel human transmembrane protein, TMEM9A and its function referred to the BC-related inflammatory response were needed to further explore. There is abundant room for further progress in determining the role of TMEM9A in BC progression.
In summary, this study provided evidence for the first time that TMEM9A was significantly expressed in BC samples, and manifested that TMEM9A played an important role in the cell proliferation, migration, and apoptosis of BC cells. Overall, by regulating the Wnt/β-catenin signaling pathway, TMEM9A exerts the tumor-promoting properties on the development of BC cells, which may be useful for the therapy of BC.
The authors declare no conflict of interest.
This article contains supplementary materials.