2018 Volume 66 Issue 3 Pages 286-294
2018 Volume 66 Issue 3 Pages 286-294
In this study, we report the identification of potent pyrimidoindazoles as phosphodiesterase10A (PDE10A) inhibitors by using the method of fragment-based drug discovery (FBDD). The pyrazolopyridine derivative 2 was found to be a fragment hit compound which could occupy a part of the binding site of PDE10A enzyme by using the method of the X-ray co-crystal structure analysis. On the basis of the crystal structure of compound 2 and PDE10A protein, a number of compounds were synthesized and evaluated, by means of structure–activity relationship (SAR) studies, which culminated in the discovery of a novel pyrimidoindazole derivative 13 having good physicochemical properties.
Schizophrenia is known as a chronic debilitating disorder affecting the psychic and motor functions of the brain1) that is characterized by a combination of positive symptoms, negative symptoms and cognitive impairments.2) Current anti-psychotics are marginally effective for treating the positive symptoms but less effective against negative symptoms and cognitive impairments. Therefore, novel type of drugs which could manage negative symptoms and cognitive impairments would represent a significant advance in schizophrenia treatment.
The phosphodiesterases (PDEs) consist of an 11-membered family of enzymes that catalyze the hydrolysis of secondary signal messengers, such as cAMP and cyclic guanosine monophosphate (cGMP). PDE10A is a dual substrate PDE that hydrolyzes both cAMP and cGMP. PDE10A is highly expressed in the brain, particularly in the medium spiny neurons of the striatum. The distribution conserves across mammalian species.3–5) Inhibition of PDE10A is considered effective in treating schizophrenia and a range of neurological, psychotic, anxiety, and movement disorders which could be benefit from increasing levels of cAMP and cGMP in neurons. Therefore, PDE10A inhibitors might prove useful in treating neurological and psychiatric disorders.6)
At present, several PDE10A inhibitors are undergoing clinical investigation7) and a large number of studies of PDE10A inhibitors have been reported.8–16) Preclinical evidence suggests that these inhibitors might demonstrate anti-psychotic, pro-cognitive, and anti-negative symptom efficacy.17–21)
We have already reported a potent PDE10A inhibitor, compound 1 (Fig. 1), as the result of the structure–activity relationship (SAR) studies on the basis of the high-throughput screening (HTS) hit compound,22) however, 1 was found to have some issues such as a metabolic instability and the moderate inhibitory activity towards CYP3A4 enzyme. For the purpose of finding other lead compounds with good physicochemical properties as clinical candidates, the method of fragment-based drug discovery (FBDD) was chosen as a novel approach.

a) Residual activities of human liver microsome (HLM) for metabolism of midazolam in the presence of test compounds were determined following pre-incubation for 30 min.
Fragment hit makes a small number of high-efficiency interactions to one of the binding pockets within the active site.23) Once fragment binders have been identified in the binding site, the objective in drug discovery is then to design potent inhibitors by using a range of strategies including fragment emerging, linking or growing.24–26) In order to make further highly efficient interactions in the active site with retaining the key interactions from the fragment hit, the hit compound has been evolved into neighboring binding pockets to produce compact and potent lead compounds. Since crystal structure of PDE10A enzyme was revealed, co-crystal structure analyses of the enzyme with compounds should be useful to design some lead compounds having desired physicochemical properties. Herein, we describe our synthetic studies on optimization from the fragment-hit compound to achieve creating potent PDE10A inhibitors by using the method of X-ray crystal structure analysis.
The synthesis of target compounds is shown in Charts 1 and 2. Synthesis of pyrazolo[1,5-a]pyrimidin-7-ol derivatives 3–12 is shown in Chart 1. β-Keto esterification of corresponding carboxylic acids 19 in the presence of monoethyl potassium malonate, 1,1′-carbonyldiimidazole (CDI) and MgCl2 followed by condensation and cyclization with 20 in AcOH gave 3–12.

Reagents and conditions: (a) Monoethyl potassium malonate, CDI, MgCl2, NEt3, THF; (b) 20, AcOH, 1,4-dioxane, 11–76% (2 steps).

Reagents and conditions: (a) NaOH aq.; (b) acetylacetone, AcOH, EtOH, 68% (2 steps); (c) monoethyl potassium malonate, CDI, MgCl2, NEt3, THF; (d) 3-aminoindazole, AcOH, 1,4-dioxane, 11% (2 steps); (e) POCl3, 38%; (f) methylamine, THF, 97%; (g) sodium methoxide, MeOH, 82%; (h) trimethylboroxine Pd(dppf)Cl2·CH2Cl2, K2CO3, 1,4-dioxane, 34%; (i) KCN, DABCO, 1,4-dioxane, H2O, 37%.
The pyrimido[1,2-b]indazole derivatives 13–18 were synthesized as depicted in Chart 2. Cyclocondensation of aminoguanidine hydrochloride 21 with succinic anhydride 22 gave a triazolopyrimidine intermediate, followed by condensation with acetylacetone to obtain carboxylic acid 23. Pyrimido[1,2-b]indazol-4-ol derivative 13 was prepared from 23 and 3-aminoindazole in a manner similar to that described in Chart 1. Chlorination of 13 with POCl3 gave 17, which was followed by displacement of methylamine, methanol or KCN yielded 14, 15 or 18, respectively. A Suzuki–Miyaura reaction of 17 with trimethylboroxine gave the methyl analogue 16.
The analyses of the X-ray co-crystal structures of PDE10A enzyme with fragment compounds from in-house compounds’ library revealed that 7-(4-chlorophenyl)-2-methylpyrazolo[1,5-a]pyrimidine (2) occupied a part of the binding site, showed good ligand efficiency (percent efficiency index (PEI)27)=0.39), meaning that this compound should be a good fragment hit (Figs. 2, 3). The pyrazolopyrimidine ring of 2 was found to occupy the active site of the PDE10A and the 4-nitrogen of pyrazolopyrimidine accepted a hydrogen bond from Gln726. The X-ray crystal structure also disclosed that 2 does not interact with Tyr693 which was found to be important to increase PDE10A selectivity in previous studies.18,28) For the purpose of preparing compounds having the interaction with Tyr693, fragment growing approach was adopted and introduction of heteroaromatics into 5-position of pyrazolopyrimidine ring was performed. In the course of preparing compounds capable of interacting with the Tyr693, the molecular weight of the compound would be considered to be increased which should cause the increase in the lipophilicity of the compounds. On the other hand, 4-chlorophenyl part was solvent exposed and was seemed not to exhibit any appreciable contribution to potency of the substrate from the X-ray co-crystal structure of PDE10A enzyme with compound 2, so that hydroxyl group was chosen for the substitution on the 7-position of pyrazolopyrimidine ring instead of 4-chlorophenyl group to reduce lipophilicity and molecular weight of the molecule. In addtion, the replacement with hydroxyl group is beneficial for quickly optimization of Tyr693 binders because of synthetic tractability for combinatorial synthesis.


The in vitro PDE10A inhibitory potency of all synthesized compounds was assessed by measuring the quantity of cAMP via the Homogeneous Time-Resolved Fluorescence (HTRF) detection method by using human PDE10A enzyme and the inhibitory activities were shown as the inhibitory ratio at the concentration of 4 µM or evaluation of IC50 about each compound.
Initial attempts at accessing the selectivity pocket was introduction of various hetero-aromatic rings containing a nitrogen atom expected to interact with Tyr693 (Table 1). In order to find a potent scaffold efficiently, we used combinatorial synthesis. Compound 3 which has the ethylene linker between quinoline and pyrazolopyrimidine showed strong inhibitory activity. Compound 4 bearing 2-quinoline substituent directly and compound 5 having methyl thioether as the linker instead of ethylene in compound 3 showed less inhibitory activity than 3, suggesting that ethylene linker brought suitable position for the quinoline nitrogen atom to interact with Tyr693. Pyridyl ethyl derivative 6 brought less potency29) than quinoline 3, indicating that phenylene part of quinoline ring would be important for the PDE10A inhibitory activity. Triazolopyrimidine 7, imidazopyridine 8, imidazopyrimidine 9 and benzimidazole 10 all showed moderate to strong inhibitory activities against PDE10A. Among them, triazolopyrimidine 7 was chosen for the further modification to improve the inhibitory potency because compound 7 showed inhibitory activity as strong as compound 3 and was found to be more hydrophilic than compound 3 (log D7.4 values of 0.3 and 2.5, respectively).
 |
X-ray co-crystal structure of triazolopyrimidine 7 and PDE10A enzyme revealed that the triazolopyrimidine unit could fill in the selectivity pocket of PDE10A enzyme as we expected, and the 4-nitrogen on triazoropyrimidine ring of 7 formed a hydrogen bond with Tyr693 (Fig. 4).

We next investigated the effect of introducing substituents on the triazolopyrimidine core and the pyrazolopyrimidine core of compound 7, respectively (Table 2). 5,7-Dimethyltriazolopyrimidine 11 showed inhibitory activity (IC50=44 nM) indicating that introduction of substituents on 5,7-position of triazolopyrimidine ring should be sterically acceptable. In contrast, 6-methyltriazolopyrimidine 12 showed weaker inhibitory activity than compound 11, suggesting that 5,7-position was favorable than 6-position. Pyrimidoindazole 13 surprisingly provided a 20-fold superior inhibitory potency in comparison with compound 11. As the catalytic pocket of PDE10A enzyme was predicted to be well occupied by compound 13, we proceeded further investigation on the basis of 13.
 |
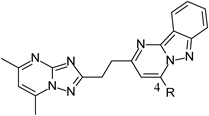
Table 3 shows the results of replacing the hydroxyl group on 4-position of the pyrimidoindazole ring with various substituents. The methyl amino derivative 14 showed strong inhibitory activity as compound 13. Although replacement with methoxy (15) or methyl (16) group caused decrease in inhibitory activity (IC50 values of 30, 64 nM, respectively), the decrease was not extreme. Replacement with chlorine (17) or cyano group (18) resulted in a much more significant decrease in PDE10A inhibitory activity (IC50 values of 221, 606 nM, respectively). The reason why hydroxyl group was the most potent as the 4-substituent was not clear unfortunately, but it was speculated that electron donating ability of the 4-substituent of the pyrimidoindazole could be important for the potent PDE10A inhibitory activity. As a result, we identified hydroxyl derivative 13 was the most potent PDE10A inhibitor.
 | ||
|---|---|---|
| Compounds | R | PDE10A IC50 (nM) |
| 13 | OH | 2.0 |
| 14 | NHMe | 7.8 |
| 15 | OMe | 30 |
| 16 | Me | 64 |
| 17 | Cl | 221 |
| 18 | CN | 606 |
The strong potency of compound 13 as a PDE10A inhibitor prompted us to evaluate its absorption, distribution, metabolism, and excretion (ADME) properties (Fig. 5). Compound 13 showed no metabolical depletion under human and mice liver microsomes condition. In addition, the inhibitory activities of compound 13 against CYP enzymes, CYP1A2, 2C8, 2C9, 2C19, 2D6 and 3A4, were found to be all weak, and P-glycoprotein (P-gp) liability of compound 13 was proved to be low.

a: Residual activities of HLM for metabolism of each substrate in the presence of test compounds were determined following pre-incubation for 30 min. b: Intrinsic clearance. c: No depletion in this condition.
The pyrazoropyrimidine derivative 13 showed strong inhibitory activity against PDE10A (IC50 value of 2.0 nM), good physicochemical properties and good metabolical stability.
Optimization of compounds guided by X-ray co-crystal structure from fragment hit 2 and PDE10A enzyme was held on for finding PDE10A inhibitors with improved physicochemical and pharmacokinetic profiles. Being able to obtain compound 7 with good inhibitory activity, we moved on to optimization of the substituents. As a result, the pyrimidoindazole derivative 13 which had potent PDE10A inhibitory activity, high metabolic stability and no significant CYPs inhibitory activity was obtained. Further investigation of PDE10A inhibitors on the basis of compound 13 as the lead compound will be described in the next report.
1H-NMR spectra were recorded on a Varian 400-MR and BRUKER AV-III HD500, and chemical shifts were expressed as δ (ppm) values with tetramethylsilane as an internal reference (s=singlet, d=doublet, t=triplet, m=multiplet, dd=double doublet, dt=double triplet, ddd=double double doublet, and br=broad peak). MS were recorded on a Waters ultra performance liquid chromatography (UPLC)/SQD and Waters Acquity UPLC/ZQ. Electrospray ionization (ESI) positive high-resolution (HR)-MS were obtained using a Thermo EXACTIVE-Plus Waters LCT Premier.
2-Methyl-5-[2-(quinolin-2-yl)ethyl]pyrazolo[1,5-a]pyrimidin-7-ol (3)A mixture of ethyl potassium malonate (1019 mg, 6.00 mmol), magnesium chloride (571 mg, 6.00 mmol) and triethylamine (1070 µL, 7.68 mmol) in tetrahydrofuran (THF) (24 mL) was stirred at room temperature for 2.5 h. One milliliter of the reaction mixture was added to another reaction mixture of 3-(quinoline-2-yl)propanoic acid (17.3 mg, 0.100 mmol) and 1,1′-carbonyldiimidazole (CDI; 24.3 mg, 0.150 mmol) in THF (0.2 mL) which had been stirred at 50°C for 2 h. The combined reaction mixture was stirred at 50°C for 15 h. After cooling at room temperature, to the reaction mixture were added 1 M HCl aqueous solution (0.5 mL), CHCl3 and saturated NaHCO3 aqueous solution (0.5 mL). The mixture was through the phase separator. The organic layer was concentrated in vacuo. To the residue was added 3-methyl-1H-pyrazole-5-amine (9.70 mg, 0.100 mmol) in 1,4-dioxane (0.100 mL) and AcOH (5.0 µL, 87 µmol), and the mixture was stirred at 90°C for overnight. After cooling at room temperature, the mixture was concentrated in vacuo. The residue was purified by preparative HPLC (Waters SunFire™ Column, C18, 5 µm, 19×100 mm, 10–95% MeOH in 0.1% (v/v) formic acid aqueous solution) to give 3 (5.4 mg, 17%) as a pale brown solid. 1H-NMR (DMSO-d6) δ: 2.27 (s, 3H), 3.09 (dd, 2H, J=9.0, 6.8 Hz), 3.32–3.36 (m, 2H), 5.57 (d, 1H, J=1.4 Hz), 5.95 (s, 1H), 7.49 (d, 1H, J=8.4 Hz), 7.56 (ddd, 1H, J=8.0, 6.9, 1.2 Hz), 7.74 (ddd, 1H, J=8.4, 6.9, 1.5 Hz), 7.93–7.96 (m, 2H), 8.30 (d, 1H, J=8.3 Hz), 12.23 (s, 1H); MS (ESI) m/z 305 [M+H]+; HR-MS (ES+) Calcd for C18H17ON4 [M+H]+ 305.1397; Found, 305.1398.
2-Methyl-5-(quinolin-2-yl)pyrazolo[1,5-a]pyrimidin-7-ol (4)Compound 4 was prepared from quinaldic acid in a manner similar to that described for compound 3, with a yield of 12%. 1H-NMR (DMSO-d6) δ: 2.34 (s, 3H), 6.22 (s, 1H), 6.72 (s, 1H), 7.75 (ddd, 1H, J=8.1, 7.0, 0.99 Hz), 7.92 (ddd, 1H, J=8.4, 6.9, 1.4 Hz), 8.12 (d, 1H, J=7.5 Hz), 8.24 (d, 1H, J=8.4 Hz), 8.37 (d, 1H, J=8.8 Hz), 8.62 (d, 1H, J=8.8 Hz), 12.53 (s, 1H); MS (ESI) m/z 277 [M+H]+; HR-MS (ES+) Calcd for C16H13ON4 [M+H]+ 277.1084; Found, 277.1084.
2-Methyl-5-[(quinolin-2-ylsulfanyl)methyl]pyrazolo[1,5-a]pyrimidin-7-ol (5)Compound 5 was prepared from (quinolin-2-ylsulfanyl)acetic acid in a manner similar to that described for compound 3, with a yield of 27%. 1H-NMR (DMSO-d6) δ: 2.26 (s, 3H), 4.52 (s, 2H), 5.84 (s, 1H), 6.00 (s, 1H), 7.47 (d, 1H, J=8.6 Hz), 7.54 (ddd, 1H, J=8.0, 7.0, 1.1 Hz), 7.76 (ddd, 1H, J=8.4, 7.0, 1.4 Hz), 7.90–7.98 (m, 2H), 8.23 (d, 1H, J=8.6 Hz), 12.43 (br s, 1H); MS (ESI) m/z 323 [M+H]+; HR-MS (ES+) Calcd for C17H15ON4S [M+H]+ 323.0961; Found, 323.0961.
2-Methyl-5-[2-(pyridin-2-yl)ethyl]pyrazolo[1,5-a]pyrimidin-7-ol (6)A mixture of ethyl potassium malonate (2145 mg, 12.6 mmol), magnesium chloride (1428 mg, 15.0 mmol) and triethylamine (2676 µL, 19.2 mmol) in THF (60 mL) was stirred at room temperature for 2.5 h. Half milliliter of the reaction mixture was added to another reaction mixture of 3-(pyridin-2-yl)propanoic acid (7.56 mg, 50.0 µmol) and CDI (12.2 mg, 75 µmol) in THF (0.2 mL) which had been stirred at 50°C for 2 h. The combined reaction mixture was stirred at 50°C for overnight. After cooling at room temperature, to the reaction mixture were added 1 M HCl aqueous solution (0.5 mL), CHCl3 (2.5 mL) and saturated NaHCO3 aqueous solution (0.5 mL). The mixture was through the phase separator. The organic layer was concentrated in vacuo. To the residue was added 3-methyl-1H-pyrazole-5-amine (4.90 mg, 50 µmol) in 1,4-dioxane (0.100 mL) and AcOH (5.0 µL, 87 µmol), and the mixture was stirred at 90°C for overnight. After cooling at room temperature, the mixture was concentrated in vacuo. The residue was purified by preparative HPLC (Waters SunFire™ Column, C18, 5 µm, 19×100 mm, 10–95% MeOH in 0.1% (v/v) formic acid aqueous solution) to give 6 (5.3 mg, 42%). MS (ESI) m/z 255 [M+H]+.
2-Methyl-5-[2-([1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]pyrazolo[1,5-a]pyrimidin-7-ol (7)Compound 7 was prepared from 3-([1,2,4]triazolo[1,5-a]pyrimidine-2-yl)propanoic acid in a manner similar to that described for compound 3, with a yield of 40% as a pale brown solid. 1H-NMR (DMSO-d6) δ: 2.27 (s, 3H), 3.05–3.13 (m, 2H), 3.23–3.30 (m, 2H), 5.57 (s, 1H), 5.94 (s, 1H), 7.31 (dd, 1H, J=6.8, 4.3 Hz), 8.83 (dd, 2H, J=4.3, 1.9 Hz), 9.33 (dd, 1H, J=6.8, 1.9 Hz), 12.20 (s, 1H); MS (ESI) m/z 296 [M+H]+; HR-MS (ES+) Calcd for C14H14ON7 [M+H]+ 296.1254; Found, 296.1257.
5-[2-(Imidazo[1,2-a]pyridin-2-yl)ethyl]-2-methylpyrazolo[1,5-a]pyrimidin-7-ol (8)Compound 8 was prepared from 3-(imidazo[1,2-a]pyridin-2-yl)propanoic acid in a manner similar to that described for compound 6, with a yield of 30%. MS (ESI) m/z 294 [M+H]+.
5-[2-(Imidazo[1,2-a]pyrimidin-2-yl)ethyl]-2-methylpyrazolo[1,5-a]pyrimidin-7-ol (9)Compound 9 was prepared from 3-(imidazo[1,2-a]pyrimidin-2-yl)propanoic acid in a manner similar to that described for compound 6, with a yield of 10%. MS (ESI) m/z 295 [M+H]+.
5-[2-(1H-Benzimidazol-2-yl)ethyl]-2-methylpyrazolo[1,5-a]pyrimidin-7-ol (10)Compound 10 was prepared from 3-(1H-benzimidazole-2-yl)propanoic acid in a manner similar to that described for compound 3, with a yield of 27%. 1H-NMR (DMSO-d6) δ: 2.27 (s, 3H), 3.09 (t, 2H, J=7.5 Hz), 3.23 (t, 2H, J=7.3 Hz), 5.54 (s, 1H), 5.95 (s, 1H), 7.08–7.16 (m, 2H), 7.43–7.53 (m, 2H), 12.30 (br s, 2H); MS (ESI) m/z 294 [M+H]+; HR-MS (ES+) Calcd for C16H16ON5 [M+H]+ 294.1349; Found, 249.1347.
3-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)propanoic Acid (23)A mixture of aminoguanidine hydrochloride 21 (25.0 g, 226 mmol) and succinic anhydride 22 (25.0 g, 250 mmol) was stirred at 190°C for 1 h. After cooling at room temperature, to the reaction mixture were added sodium hydroxide (27.0 g, 675 mmol) and H2O (67 mL). The mixture was stirred at 120°C for 1.5 h. After cooling, to the reaction mixture were added conc. HCl aqueous solution (37 mL) and H2O (25 mL) at room temperature, and the mixture was stood overnight. The resulting precipitate under ice-bath cooling was collected by filtration. To the residue were added EtOH (400 mL), acetylacetone (20.0 mL, 194 mmol) and AcOH (2.00 mL, 35.0 mmol), and the mixture was stirred under reflux for 7 h. After cooling at room temperature, to the mixture were added H2O (30 mL) and sodium hydroxide (10.0 g, 250 mmol), and the mixture was stirred at room temperature for 2 h. The mixture was filtered and the filtrate was concentrated in vacuo lightly. To the solution was added conc. HCl aqueous solution (20 mL) and the mixture was cooled under ice-bath. The resulting precipitate was collected by filtration to give 23 (33.7 g, 68%) as a colorless solid. 1H-NMR (DMSO-d6) δ: 2.55 (s, 3H), 2.69 (d, 3H, J=0.9 Hz), 2.76 (t, 2H, J=7.3 Hz), 3.05 (t, 2H, J=7.4 Hz), 7.10 (d, 1H, J=0.9 Hz), 12.20 (br s, 1H); MS (ESI) m/z 221 [M+H]+.
5-[2-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]-2-methylpyrazolo[1,5-a]pyrimidin-7-ol (11)To a mixture of 23 (661 mg, 3.00 mmol) in THF (10 mL) was added CDI (584 mg, 3.60 mmol), and the mixture was stirred at 50°C for 1 h. To the reaction mixture were added ethyl potassium malonate (1.02 g, 6.00 mmol), magnesium chloride (571 mg, 6.00 mmol), triethylamine (1.04 mL, 7.50 mmol), and the mixture was stirred at 50°C for 12 h. After cooling at room temperature, to the reaction mixture was added 1 M HCl aqueous solution, and the mixture was stirred at ambient temperature for 1 h. The mixture was extracted with EtOAc. The organic layer was washed with brine, dried over anhydrous Na2SO4, filtered and concentrated in vacuo. To the residue were added 1,4-dioxane (10 mL), 5-methyl-1H-pyrazol-3-amine (200 mg, 2.06 mmol) and AcOH (1.18 mL, 20.6 mmol), and the mixture was stirred at 90°C for 12 h. After cooling at room temperature, the mixture was diluted with Et2O, and the resulting precipitate was collected by filtration to give 11 (614 mg, 92%) as a beige solid. 1H-NMR (DMSO-d6) δ: 2.27 (s, 3H), 2.56 (s, 3H), 2.68 (d, 3H, J=1.0 Hz), 3.01–3.11 (m, 2H), 3.20–3.28 (m, 2H), 5.57 (s, 1H), 5.94 (s, 1H), 7.12 (d, 1H, J=0.8 Hz), 12.20 (br-s, 1H); MS (ESI) m/z 324 [M+H]+; HR-MS (ES+) Calcd for C16H18ON7 [M+H]+ 324.1567; Found, 324.1569.
2-Methyl-5-[2-(6-methyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]pyrazolo[1,5-a]pyrimidin-7-ol (12)To a mixture of 3-(6-methyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)propanoic acid (619 mg, 3.00 mmol) in THF (10 mL) was added CDI (584 mg, 3.60 mmol), and the mixture was stirred at 50°C for 1 h. To the reaction mixture were added ethyl potassium malonate (1.02 g, 6.00 mmol), magnesium chloride (571 mg, 6.00 mmol), triethylamine (1.04 mL, 7.50 mmol), and the mixture was stirred at 50°C for 12 h. After cooling at room temperature, to the reaction mixture was added 1 M HCl aqueous solution, and the mixture was stirred at ambient temperature for 1 h. The mixture was extracted with EtOAc. The organic layer was washed with brine, dried over anhydrous Na2SO4, filtered and concentrated in vacuo. To the residue were added 1,4-dioxane (10 mL), 5-methyl-1H-pyrazol-3-amine (200 mg, 2.06 mmol) and AcOH (1.18 mL, 20.6 mmol), and stirred at 90°C for 12 h. After cooling at room temperature, the mixture was diluted with Et2O, and the resulting precipitate was collected by filtration to give 12 (487 mg, 76%) as a beige solid. 1H-NMR (DMSO-d6) δ: 2.26 (s, 3H), 2.36 (d, 3H, J=0.8 Hz), 3.01–3.11 (m, 2H), 3.20–3.29 (m, 2H), 5.55 (s, 1H), 5.93 (s, 1H), 8.73 (d, 1H, J=2.4 Hz), 9.17 (dd, 1H, J=2.3, 1.1 Hz), 12.20 (s, 1H); MS (ESI) m/z 310 [M+H]+; HR-MS (ES+) Calcd for C15H16ON7 [M+H]+ 310.1411; Found, 310.1414.
2-[2-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]pyrimido[1,2-b]indazol-4-ol (13)To a mixture of 23 (2.20 g, 10.0 mmol) in THF (33 mL) was added CDI (1.95 g, 12.0 mmol), and the mixture was stirred at 50°C for 1 h. To the reaction mixture were added ethyl potassium malonate (3.40 g, 20.0 mmol), magnesium chloride (1.90 g, 20.0 mmol), triethylamine (3.48 mL, 25.0 mmol), and the mixture was stirred at 50°C for 12 h. After cooling at room temperature, to the reaction mixture was added 1 M HCl aqueous solution (50 mL), and the mixture was stirred at ambient temperature for 1 h. The mixture was extracted with EtOAc. The organic layer was washed with brine, dried over anhydrous Na2SO4, filtered and concentrated in vacuo. To a half of the residue were added 1,4-dioxane (15 mL), 1H-indazol-3-amine (610 mg, 4.58 mmol) and AcOH (4 mL), and stirred at 95°C for 18 h. After cooling at room temperature, the mixture was concentrated in vacuo. The residue was diluted with EtOAc and saturated NaHCO3 aqueous solution, and the resulting precipitate was collected by filtration to give 13 (194 mg, 11%) as a beige solid. 1H-NMR (DMSO-d6) δ: 2.56 (s, 3H), 2.70 (d, 3H, J=0.7 Hz), 3.03–3.13 (m, 2H), 3.21–3.27 (m, 2H), 5.76 (s, 1H), 6.82 (ddd, 1H, J=7.9, 6.8, 0.8 Hz), 7.09 (d, 1H, J=0.9 Hz), 7.24–7.33 (m, 1H), 7.44 (d, 1H, J=8.6 Hz), 7.90 (dt, 1H, J=8.2, 1.0 Hz); MS (ESI) m/z 360 [M+H]+; HR-MS (ES+) Calcd for C19H18ON7 [M+H]+ 360.1567; Found, 360.1568.
4-Chloro-2-[2-(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]pyrimido[1,2-b]indazole (17)A mixture of 13 (280 mg, 0.779 mmol) and phosphorous oxychloride (3.35 g, 21.8 mmol) was stirred at 100°C for 1 d. After cooling at room temperature, the mixture was concentrated in vacuo. The residue was diluted with saturated NaHCO3 aqueous solution and extracted with CHCl3. The organic layer was washed with brine, dried over anhydrous MgSO4, filtered, and concentrated in vacuo. The residue was purified by flash column chromatography (silica gel, 0–3% MeOH in CHCl3) to give 17 (112 mg, 38%) as a yellow solid. 1H-NMR (DMSO-d6) δ: 2.55 (s, 3H), 2.68 (d, 3H, J=0.9 Hz), 3.36–3.58 (m, 4H), 7.10 (d, 1H, J=0.9 Hz), 7.32 (ddd, 1H, J=8.3, 6.7, 0.7 Hz), 7.67 (ddd, 1H, J=8.7, 6.7, 1.1 Hz), 7.87 (dt, 1H, J=8.6, 0.9 Hz), 7.98 (s, 1H), 8.23 (dt, 1H, J=8.3, 1.0 Hz); MS (ESI) m/z 378 [M+H]+; HR-MS (ES+) Calcd for C19H17N7Cl [M+H]+ 378.1228; Found, 378.1230.
2-[2-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]-N-methylpyrimido[1,2-b]indazol-4-amine (14)To a solution of 17 (96.0 mg, 0.254 mmol) in THF (0.96 mL) was added methylamine solution (381 µL, 0.762 mmol) and the mixture was stirred at room temperature for 17 h. The mixture was concentrated in vacuo, and the residue was purified by flash column chromatography (silica gel, 0–20% MeOH in CHCl3) to give 14 (92.0 mg, 97%) as a yellow solid. 1H-NMR (DMSO-d6) δ: 2.56 (s, 3H), 2.70 (d, 3H, J=0.7 Hz), 3.04 (d, 3H, J=4.9 Hz), 3.33–3.43 (m, 4H), 6.58 (s, 1H), 7.05–7.14 (m, 2H), 7.43–7.58 (m, 1H), 7.66 (d, 1H, J=8.6 Hz), 8.05–8.13 (m, 1H), 8.14–8.23 (m, 1H); MS (ESI) m/z 373 [M+H]+; HR-MS (ES+) Calcd for C20H21N8 [M+H]+ 373.1884; Found, 373.1886.
2-[2-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]-4-methoxypyrimido[1,2-b]indazole (15)To a solution of 17 (100 mg, 0.265 mmol) in MeOH (1.00 mL) was added sodium methoxide (28.6 mg, 0.529 mmol) and the mixture was stirred at room temperature for 3 h. The reaction mixture was diluted with H2O, and the resulting precipitate was collected by filtration. The residue was purified by flash column chromatography (silica gel, 0–10% MeOH in CHCl3). The residue was washed with EtOAc to give 15 (81.0 mg, 82%) as a yellow solid. 1H-NMR (DMSO-d6) δ: 2.56 (s, 3H), 2.70 (d, 3H, J=0.9 Hz), 3.39–3.55 (m, 4H), 4.28 (s, 3H), 7.11 (d, 1H, J=0.9 Hz), 7.14–7.29 (m, 2H), 7.56 (ddd, 1H, J=8.8, 6.7, 1.1 Hz), 7.72 (dt, 1H, J=8.7, 0.9 Hz), 8.15 (dt, 1H, J=8.3, 1.0 Hz); MS (ESI) m/z 374 [M+H]+; HR-MS (ES+) Calcd for C20H20ON7 [M+H]+ 374.1724; Found, 374.1726.
2-[2-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]-4-methylpyrimido[1,2-b]indazole (16)To a stirred mixture of 17 (50.0 mg, 0.132 mmol), trimethylboroxine (49.8 mg, 0.397 mmol) and [1,1′-bis(diphenylphosphino)ferrocene]palladium(II) dichloride dichloromethane adduct (Pd(dppf)Cl2·CH2Cl2; 32.4 mg, 39.7 µmol) in 1,4-dioxane (1.00 mL) was added K2CO3 (110 mg, 0.794 mmol) under argon atmosphere, and the mixture was stirred at 90°C for 1 d. After cooling at room temperature, the mixture was diluted with water and extracted with CHCl3. The organic layer was washed with brine, dried over anhydrous MgSO4, filtered and concentrated in vacuo. The residue was purified by flash column chromatography (silica gel, 0–5% MeOH in CHCl3) to give 16 (16.0 mg, 34%) as a pale yellow solid. 1H-NMR (DMSO-d6) δ: 2.55 (s, 3H), 2.69 (d, 3H, J=0.7 Hz), 2.88 (d, 3H, J=0.7 Hz) 3.36–3.54 (m, 4H), 7.10 (d, 1H, J=0.9 Hz), 7.24 (ddd, 1H, J=8.2, 6.7, 0.8 Hz), 7.43–7.65 (m, 2H), 7.70–7.90 (m, 1H), 8.20 (dt, 1H, J=8.3, 0.9 Hz); MS (ESI) m/z 358 [M+H]+; HR-MS (ES+) Calcd for C20H20N7 [M+H]+ 358.1775; Found, 358.1776.
2-[2-(5,7-Dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]pyrimido[1,2-b]indazole-4-carbonitrile (18)A mixture of 17 (50.0 mg, 0.132 mmol), potassium cyanide (18.0 mg, 0.276 mmol) and 1,4-diazabicyclo[2.2.2]octane (DABCO; 18.0 mg, 0.160 mmol) in 1,4-dioxane (2.0 mL) and H2O (0.20 mL) was stirred at room temperature for 5 h. The residue was diluted with H2O. The resulting precipitate was collected by filtration and purified by flash column chromatography (silica gel, 0–5% MeOH in CHCl3) to give 18 (18.0 mg, 37%) as a bright red solid. 1H-NMR (DMSO-d6) δ: 2.55 (s, 3H), 2.68 (s, 3H), 3.40–3.60 (m, 4H), 7.10 (s, 1H), 7.36–7.50 (m, 1H), 7.73 (ddd, 1H, J=8.4, 7.0, 1.0 Hz), 7.94 (d, 1H, J=8.6 Hz), 8.26–8.36 (m, 1H), 8.40 (s, 1H); MS (ESI) m/z 369 [M+H]+; HR-MS (ES+) Calcd for C20H17N8 [M+H]+ 369.1571; Found, 369.1571.
Human PDE10A phosphodiesterase domain (amino acids 449–789) was cloned between the NdeI and XhoI sites of pET28a vector to express target protein as a 6×histidine (His)-tagged protein. This plasmid was transformed in Escherichia coli BL21(DE3) and overexpressed. The cells were disrupted by sonication and the supernatant was collected by centrifugation at 4°C. The target protein was precipitated from the supernatant by adding ammonium sulfate to 60% and was collected by centrifugation. The precipitate was dissolved with nickel–nitrilotriacetic acid (Ni–NTA) wash buffer (25 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole) and was applied onto an Ni–NTA column. After washing with Ni–NTA wash buffer, the bound protein was eluted with 25 mM Tris–HCl pH 8.0, 500 mM NaCl, 250 mM imidazole. Eluate was collected and the buffer exchanged with 25 mM Tris–HCl pH 8.0, 500 mM NaCl. The amino-terminal His tag of PDE10A (449–789) was removed by incubation with thrombin protease for 12 h at 4°C. After digestion, protein solution was passed through Benzamidine Sepharose 6B column (GE Healthcare, U.S.A.) and again applied onto an Ni–NTA column to remove thrombin and uncleaved His-tag protein. Further purification was performed by passing through Q Sepharose column (GE Healthcare) equilibrated with 25 mM Tris–HCl pH 8.0, 150 mM NaCl followed by Superdex 75 pg 16/60 (GE Healthcare) in 25 mM N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (HEPES)-Na pH 7.5, 100 mM NaCl.
PDE10A Inhibition Assay (for Inhibition Assay of Compounds 2–10)Inhibition of human PDE10A enzyme activity was assessed by measuring the quantity of cAMP via the Homogeneous Time-Resolved Fluorescence (HTRF) detection method. The assay was performed in 20 µL samples containing an optimal amount of the PDE10A enzyme domain, a buffer (50 mM Tris–HCl pH 8.0; 2 mM MgCl2, 0.1% BSA), 0.1 µM cAMP and various concentrations of compounds (0.03 nM to 40 µM). After compounds were mixed with the enzyme, the reaction was initiated by adding the substrate cAMP and the mixture was incubated for 30 min at room temperature with agitation. The reaction was terminated on addition of the fluorescence acceptor (cAMP labeled with the dye d2) and the fluorescence donor (anti-cAMP antibody labeled with Cryptate, Cisbio). After 60 min, the fluorescence transfer corresponding to the amount of residual cAMP was measured at lex. 320 nm, lem. 620 nm and lem. 665 nm using an Envision plate reader (PerkinElmer, Inc., U.S.A.) and signal ratio (665/620) was calculated. The ratio determined in the absence of enzyme was subtracted from all data. The obtained results were converted to inhibition percentage relative to an uninhibited control.
PDE10A Inhibition Assay (for Inhibition Assay of Compounds 11–18)Inhibitory activity of compounds on human PDE10A2 was assessed by measuring the residual amount of cAMP, substrate for PDE10A2, by the HTRF detection method (cisbio).8) The obtained results were converted to activity relative to an uninhibited control (100%) and IC50 values were calculated using Prism software (GraphPad Software, Inc., U.S.A.).
Crystallization and Structural AnalysisPDE10A crystals were obtained as previously reported.30) Briefly, crystals were precipitated by sitting drop vapor diffusion methods. The reservoir solution contains 0.05 M bis-tris propane (pH 6.0), 0.1 M magnesium sulfate, 15–18% (w/v) PEG3350. Compounds were soaked into the apo PDE10A crystal. An apo crystal was transferred to a mother liquor containing compound (5 mM final concentration) and incubated at 4°C for three days. X-ray diffraction data were collected at AR-NE3A beamline31) at the Photon Factory in the National Laboratory for High Energy Physics (KEK), Tsukuba, Japan. Diffraction data were indexed, integrated, and scaled using HKL2000.32) Crystal structures of PDE10A were determined by the molecular replacement method using AMoRe33) with 2OUN30) as a search model. An initial refinement was performed using REFMAC.34) Compounds were fitted into electron densities observed in initial Fo-Fc maps using AFITT (OpenEye Scientific Software, NM, U.S.A.). Water placements and further refinements were performed using Coot35) and REFMAC, and the final models were determined. Data collection and refinement statistics are summarized in Table 4.
| Compound | 2 | 7 |
|---|---|---|
| PDB ID | 5XUJ | 5XUI |
| Space group | P212121 | P212121 |
| a (Å) | 50.3 | 49.3 |
| b (Å) | 81.3 | 81.4 |
| c (Å) | 157.6 | 158.7 |
| α (°) | 90 | 90 |
| β (°) | 90 | 90 |
| γ (°) | 90 | 90 |
| Resolution (Å) | 50.0–2.44 (2.48–2.44) | 50.0–2.77 (2.82–2.77) |
| Multiplicity | 4.8 (5.0) | 5.5 (5.1) |
| Average I/σ(I) | 31.7 (5.8) | 25.2 (4.1) |
| Rmergea) (%) | 7.3 (35.6) | 8.7 (41.9) |
| No. of reflections | 21730 (1567) | 15497 (1103) |
| Completeness (%) | 92.0 (91.7) | 96.6 (98.2) |
| Rworkb) (%) | 19.8 | 18.5 |
| Rfreec) (%) | 28.0 | 27.2 |
| Average B factor (Å2) | 52.4 | 57.6 |
| RMSD bond length (Å) | 0.011 | 0.010 |
| RMSD bond angle (°) | 1.569 | 1.461 |
Values for the outer shell are given in parentheses. a) Rmerge=∑hkl ∑i|Ii−I|/∑hkl ∑iIi, where Ii is the intensity of an individual reflection and I is the mean intensity obtained from multiple observations of symmetry related reflections. b) Rwork=∑hkl||Fobs|−|Fcalc||/∑hkl|Fobs|. c) Five percent randomly omitted reflections were used for Rfree.
Pooled mouse or human liver microsomes (Xenotech LLC., U.S.A.) were diluted in 100 mM KH2PO4/K2HPO4 buffer (pH 7.4) containing 0.1 mM ethylenediaminetetraacetic acid (EDTA). The incubation mixtures (270 µL total volume), which contained 0.2 mg/mL of microsomal proteins, and 0.2 µM of substrates were pre-incubated for approximately 15 min at 37°C. Reactions were initiated by the addition of 1 mM reduced nicotinamide adenine dinucleotide phosphate (NADPH) (30 µL). After the appropriate incubation time (0, 15, 30, 45 min), 40 µL of incubation mixture was transferred into 80% acetonitrile containing internal standard (200 nM methyltestosterone, 250 µL), stood at 4°C for 20 min, and centrifuged for 20 min at 2800 rpm. The supernatant (200 µL) was prepared and analyzed via LC-MS/MS with UPLC system (Waters) and Xevo TQ (Waters). The in vitro intrinsic clearance (CLint, vitro) was calculated using Eq. 1, which is based on the time course of the residual ratio of the compounds.36)
 | (1) |
Time-dependent inhibition assay for CYP activity was performed in two steps, a pre-incubation step where the test compound was incubated with human liver microsomes and the secondary incubation period where specific substrates were added to the preincubate to measure residual CYP activity. Specific metabolites were used to monitor the CYP activities.
Each test compound (5 µM) was pre-incubated with human liver microsomes (0.1 mg/mL) and NADPH (1.0 mM) at 37°C. The pre-incubation times used were 0 and 30 min. Following the pre-incubation step, each compound was co-incubated with substrates at 37°C for 20 min. At the end of the incubation, the reaction was terminated by the addition of aqueous solution containing 80% acetonitrile. The concentration of metabolites was determined by LC-MS analysis. The inhibition of CYP activities was assessed by comparing the amount of metabolites formed in the presence of single concentration of inhibitor to the amount of metabolites formed in the solvent control. In each study, a CYP potent and specific inhibitors were used as positive control.
 | (2) |
The substrates of CYP1A2, 2C8, 2C9, 2C19, 2D6 and 3A4 were phenacetin (20 µM), amodiaquine (0.1 µM), diclofenac (10 µM), S-mephenytoin (30 µM), dextromethorphan (7.0 µM) and midazolam (1.5 µM), respectively.
Transcellular Transport Study in LLC-PK1-Multiple Drug Resistance 1 (MDR1) CellsWild type or MDR1-expressing LLC-PK1 cells (LLC-PK1-WT or LLC-PK1-MDR1, respectively) cultured for 5 d on a Millicell-96 Cell Culture Insert Plate (Millipore) were pre-incubated with transport buffer (HBSS, pH 7.4, for the apical and basolateral sides) for 1 h. After aspiration of the transport buffer, the donor solution (transport buffer (0.5% dimethyl sulfoxide (DMSO)) containing the test compound (1 µM) and Texas Red (1 µM)) was added to the apical or basolateral side for the influx or efflux transport study, respectively, and the receiver solution (transport buffer (0.5% DMSO)) was added to the opposite side. After incubation for 3 h, the test compound in both sides was analyzed by LC/MS/MS and the apparent permeability was determined. Efflux ratio (ER) was calculated by dividing the apparent permeability in the direction from the basolateral to the apical side by that in the opposite direction. Net efflux ratio (NER) was the ratio of ER of LLC-PK1-MDR1 to LLC-PK1-WT. Texas Red was used for the estimation of the apparent permeability via para cellular transport.
The authors wish to thank to Dr. Takeshi Shimada for performing pharmacological evaluations, Mr. Hiroyuki Moriguchi for his helpful support in preparing this manuscript, and the staff of Astellas Research Technologies Co., Ltd. for conducting the CYP inhibition screening, metabolic clearance assay, partition coefficient assay, elemental analysis, and spectral measurements.
All authors were employees of Astellas Pharma Inc. when the study was conducted and have no further conflicts of interest to declare.