2019 Volume 67 Issue 11 Pages 1242-1247
2019 Volume 67 Issue 11 Pages 1242-1247
Polygalaxanthone III, a xanthone glycoside that is a major constituent of “Polygala Root” (Polygala tenuifolia roots, Onji in the Japanese Pharmacopoeia), has been used as a standard in the quality control of crude drugs. However, we previously noted differences in the chromatographic properties of one of three samples of polygalaxanthone III. Therefore, standardization of the standard itself is extremely important. The structures of three standard samples commercially available as polygalaxanthone III were characterized by LC/MS and NMR. LC/MS analysis revealed that two molecular types exist. Both types are chromatographically separable but have an identical mass number with distinguishable MS/MS spectra. One dimensional (1D)-NMR analyses demonstrated that both had the same xanthone moiety and heteronuclear multiple bond correlation (HMBC) analyses revealed that they are structural isomers at the connecting position of glucose to apiose 1-position. Consequently, the isomers were identified as polygalaxanthone III and its regioisomer, polygalaxanthone XI. Based on the findings, we recommend using the LC-MS/MS detection method, which discriminates polygalaxanthone III and XI, to confirm the quality of the standard.
Onji (Polygala Root, POLYGALAE RADIX) is a herbal medicine obtained by drying the roots or root bark of Polygala tenuifolia Willdenow (Polygalaceae).1) It is used as a herbal raw material in the preparation of crude drug products for the improvement of memory in middle aged and older individuals and is a component of Kampo formulae such as Kamiuntanto and Kihito, both of which are used for insomnia, neurosis, and mental instability.2) Considering the efficacy of Onji extract as a single herbal medicine, OTC Onji products have been launched by several pharmaceutical companies in recent years. As a result, it has become necessary to establish a method for their quality evaluation using marker compounds.3)
Characteristically, various xanthone glycosides are included in the roots of P. tenuifolia. Polygalaxanthone (PGX) III was first isolated and identified from P. tenuifolia roots in 1994, and its structure was estimated as 4-C-[β-D-apiofuranosyl-(1→6)-β-D-glycopyranosyl]-1,3,6-trihydroxy-7-methoxyxanthone.4) In 1999, the structure of PGX III was revised as 2-C-[β-D-apiofuranosyl-(1→6)-β-D-glucopyranosyl]-1,3,6-trihydroxy-7-methoxyxanthone.5) Some isomers of PGX III were also identified from the roots of P. tenuifolia.6) Since the PGX III molecule has been used as a standard substance for the quality control of crude drugs in the Pharmacopoeia of the People’s Republic of China,7) it has been commercially available from several manufacturers. Using such a standard substance, Nishihara et al. reported a quantification method for PGX III in Onji.8,9)
During our successive studies to establish a quality control method for single herbal extracts,10) we found that two of the purchased PGX III samples showed the same retention time by LC analysis, but one showed a different time. It is thus important to identify the true structure being used as the reference sample for quality control methods. Thus, we initiated their structural identification using LC-MS/MS and NMR. In this study, these detailed structures namely PGX III and PGX XI, are described.
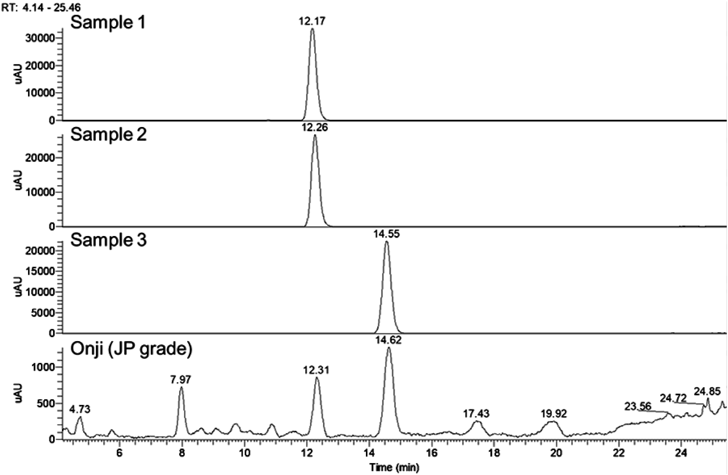
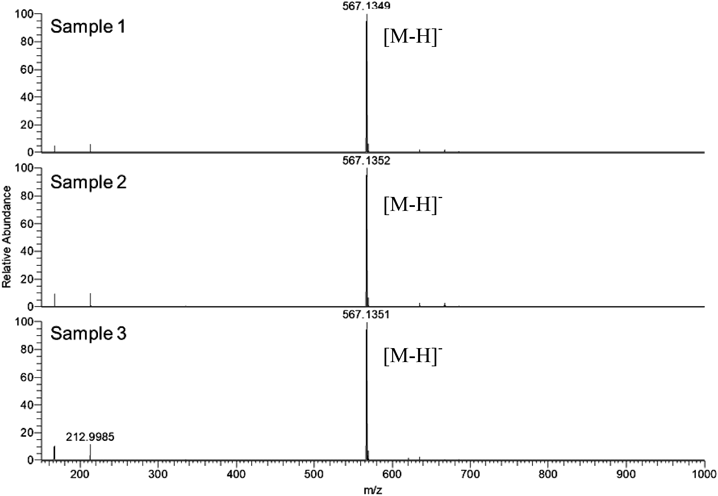
First, three commercially available PGX III standards were purchased from three different manufactures along with the crude drug Onji (Japanese Pharmacopoeia (JP) grade), and were analyzed using UHPLC/MS (Fig. 1). The retention times of samples 1 and 2 coincided with each other, but the retention time of sample 3 was different from that of the others. Comparison with the reference extract from the commercial crude drug Onji (JP grade) confirmed that both these compounds are present in Onji. LC-MS analyses showed that samples 1, 2, and 3 showed the same molecular weight based of their quasi molecular ion data (m/z 567.135 [M−H]−) and their molecular formulae were presumed as C25H28O15 (Fig. 2), which is identical to that of PGX III or its isomers including PGX VIII,6) or XI. In the electrospray ionization (ESI) (−)-MS/MS spectra, fragment ions at m/z = 315 and 345 were observed in all samples. In samples 1 and 2, fragment ions at m/z = 417 and 447 were observed characteristically but these were not detected in sample 3 (Fig. 3).

To further confirm the structure of these samples, we first measured 1H-NMR at 35°C according to a previous report,5) and found that samples 1 and 2 yielded a mixture-like spectrum; we thought that the presence of rotamers might cause this phenomenon. In another report,6) the NMR spectrum for PGX XI was measured at 70°C; when we measured the samples at the same temperature, the complexity of the spectra could not be perfectly resolved (data not shown). Therefore, we decided to analyze the 1H- and 13C-NMR spectra of samples 1, 2, and 3 at 100°C to converge the mixture of rotamers, and thus obtained good results derived from single forms.
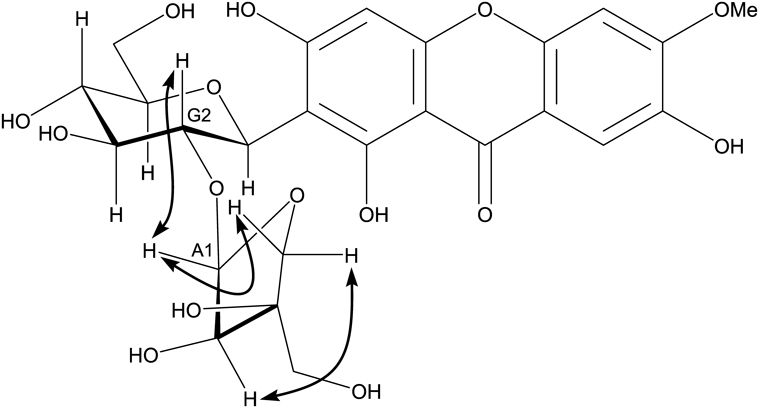
1H- and 13C-NMR data at 100°C (Figs. S1 and S2, Supplementary Materials) showed that samples 1 and 2 are identical to each other, whereas sample 3 is different from either sample 1 and 2, and that samples 1, 2, and 3 have the same aglycone moiety (1,3,6-trihydroxy-7-methoxyxanthone) with two sugars, glucose and apiose. The chemical shift of the 1-position of glucose were observed at 4.65–4.7 ppm for 1H-NMR, and 72–74 ppm for 13C-NMR, indicating that this glucose was connected to the aglycone by C–C bonds. If glucose was connected to the xanthone by C–O bonds, such as PGX V (6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-glucopyranosyl]-1,3,6-trihydroxy-7-methoxyxanthone), the chemical shift of the anomeric position of glucose would be appeared around 5.40 ppm for 1H-NMR, and 97.0 ppm for 13C-NMR.11) In addition, Jvicinal values (8–10 Hz) of endocyclic hydrogens at the glucose moieties showed that its conformer was a chair with a β-glycosidic linkage. Comparing our proton and carbon spectral data to those in previous reports, the data for samples 1 and 2 are very similar to those of PGX XI and those of the sample 3 are similar to those of PGX III except for the proton signals at positions Glc-5, 6 and Api-4, 5. Since the difference in the structure of PGX III and XI is the connecting portion of apiose to glucose, we measured the heteronuclear multiple bond correlation (HMBC) spectra of samples 1 and 3 after determining the direct C–H bond correlations by heteronuclear multiple quantum correlation (HMQC). As shown in Fig. 4, the proton signals at the Api-1 position in the spectrum of samples 1 and 3 are apparently connected to the carbon signal at the Glc-2 and Glc-6 positions, respectively. The connecting position (Api-1 to Glc-2) between apiose and glucose in sample 1 was further confirmed by one dimensional (1D)-rotating frame nuclear Overhauser effect spectroscopy (ROESY) analysis (Fig. 5). In addition, the ROESY analysis showed that the apiose had a β-glycosidic linkage. These data clearly indicate that samples 1 and 2 are PGX XI and that sample 3 is PGX III.

Recently, our UHPLC studies to establish a quality control method for Onji extract revealed a different component with a UV absorbance spectrum similar to PGX III (sample 3) at the neighboring retention time (Fig. 1). Co-elution analysis of the Onji extract and sample 1 suggested that this component was PGX XI.
Next, we tried to develop a UHPLC discrimination method between PGX III and XI without their authentic samples, as we found that indication of commercial standard products on PGX derivatives was sometimes imprecise. Because of the similarity in their UV spectra, PDA detection was not an option and we selected MS/MS detection. ESI (−)-MS spectra of PGX III and XI, and their MS/MS spectra when M − H− at m/z 567 collides are shown in Fig. 2 and Fig. 3, respectively.
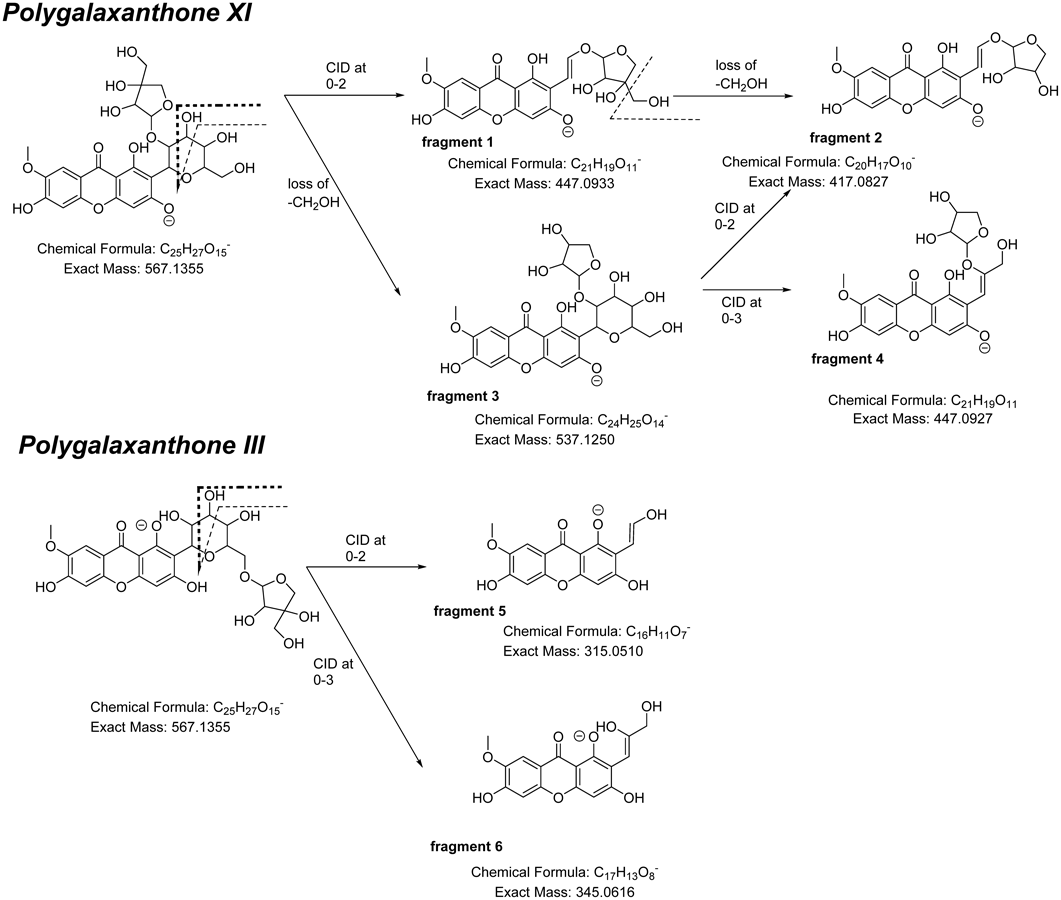
Two groups reported that in ESI (−)-LC-MS/MS, aryl-C-glucoside has a fragment in the form of collision-induced dissociation (CID) of the glycan moiety at 0–3 bond and 0–2 bond.12,13) Based on their results, the plausible fragmentations of PGX III and XI are shown in Chart 1. The m/z = 447 fragment (fragment 1) in the MS/MS spectra of samples 1 and 2 corresponded to the part of PGX XI for which positions 3 to 6 in the glucose moiety were eliminated due to cleavage at the 0–2 bond. The peak of m/z = 417 (fragment 2) was thought to be generated by elimination of the exocyclic hydroxymethyl group of the apiose moiety in fragment 1. Elimination of the hydroxymethyl group of the apiose moiety from PGX XI generated fragment 3 followed by the CID at the 0–2 or 0–3 bond, which generated fragment 2 at m/z = 417 or fragment 4 at m/z = 447, respectively. In contrast, PGX III CIDs at 0–2 bond and 0–3 bond could produce fragment 5 (m/z = 315) and fragment 6 (m/z = 345), respectively. Since the apiose moiety was connected to the glucose moiety at the 6-position in PGX III, the fragment of the C-glucosyl xanthone framework did not include an apiose moiety under the CID. On the contrary, in case of PGX XI, its apiose moiety was connected to the glucose moiety at the 2-position. Therefore, the architecture of the fragment of C-glucosyl xanthone would include the apiose moiety under the CID. Consequently, we found that MS/MS detection was a suitable method to discriminate PGX III and XI without authentic samples.
| Measured value | Literature value | ||||
|---|---|---|---|---|---|
| Sample 1a) | Sample 3a) | PGX III5, b) | PGX XI6, c) | PGX V11, c) | |
| Temperature (°C) | 100 | 100 | 35 | 70 | |
| Position | δ (ppm) | δ (ppm) | δ (ppm) | δ (ppm) | δ (ppm) |
| H-2 | 6.36 (d, 1.8) | ||||
| H-4 | 6.34 (s) | 6.35 (s) | 6.40 (s) | 6.37 (s) | 6.18 (d, 1.8) |
| H-5 | 6.86 (s) | 6.85 (s) | 6.91 (s) | 6.89 (s) | 7.31 (s) |
| H-8 | 7.47 (s) | 7.46 (s) | 7.46 (s) | 7.48 (s) | 7.44 (s) |
| MeO | 3.87 (s) | 3.87 (s) | 3.89 (s) | 3.89 (s) | 3.85 (s) |
| Glc-1 | (ax) 4.70 (d, 9.8) | (ax) 4.65 (d, 9.8) | 4.60 (d, 10) | 4.69 (d, 9.9) | 5.40 (d, 7.8) |
| Glc-2 | (ax) 4.19 (br) | (ax) 3.97 (dd, 9.8, 8.9) | 4.06 (dd, 10, 9) | na | 3.61 (t, 8.4) |
| Glc-3 | (ax) 3.42 (t, 9.0) | (ax) 3.25 (t, 8.9) | 3.32 (dd, 9, 9) | na | 3.48 (m) |
| Glc-4 | (ax) 3.24 (t, 9.0) | (ax) 3.19 (t, 8.9) | 3.22 (dd, 9, 9) | na | 3.20 (m) |
| Glc-5 | (ax) 3.20 (ddd, 9.0, 5.5, 2.5) | 3.37–3.32 (m) | 3.11 (m) | na | 3.49 (m) |
| Glc-6 | 3.68 (dd, 11.8, 2.5) | 3.85 (dd, 11.2, 1.9) | 3.35 (m) | na | 3.69 (m) |
| Glc-6 | 3.48 (dd, 11.8, 5.5) | 3.47 (dd, 11.2, 6.2) | 3.35 (m) | na | 3.45 (m) |
| Api-1 | 5.16 (d, 1.5) | 4.79 (d, 1.9) | 4.78 (d, 3) | 5.19 (s) | |
| Api-2 | 3.58 (brs) | 3.75 (d, 1.9) | 3.75 (d, 3) | na | |
| Api-4a | 2.74 (d, 9.2) | 3.57 (d, 9.4) | 3.34 (d, 11) | na | |
| Api-4b | 3.09 (d, 9.2) | 3.83 (d, 9.4) | 3.31 (d, 11) | na | |
| Api-5 | 3.15 (d, 11.2) | 3.36 (d, 11.1) | 3.86 (d, 9.5) | na | |
| Api-5 | 3.01 (d, 11.2) | 3.33 (d, 11.1) | 3.58 (d, 9.5) | na | |
| Rha-1 | 5.25 (s) | ||||
| Rha-2 | 3.30 (m) | ||||
| Rha-3 | 3.68 (m) | ||||
| Rha-4 | 3.16 (m) | ||||
| Rha-5 | 3.84 (m) | ||||
| Rha-6 | 1.13 (d, 5.7) | ||||
na: not assigned. a) Recorded in DMSO-d6 at 800 MHz. b) Recorded in DMSO-d6 at 400 MHz. c) Recorded in DMSO-d6 at 300 MHz.
| Measured value | Literature value | ||||
|---|---|---|---|---|---|
| Sample 1a) | Sample 3a) | PGX III5, b) | PGX XI6, c) | PGX V11, c) | |
| Temperature (°C) | 100 | 100 | 35 | 70 | |
| Position | δ (ppm) | δ (ppm) | δ (ppm) | δ (ppm) | δ (ppm) |
| C-1 | 162.18 | 162.27 | 161.7 | 162.7 | 162.6 |
| C-2 | 108.30 | 108.28 | 107.6 | 107.3 | 98.0 |
| C-3 | 164.42 | 164.45 | 163.8 | 163.7 | 165.1 |
| C-4 | 94.23 | 94.41 | 93.4 | 93.2 | 93.7 |
| C-4a | 157.05 | 157.00 | 156.1 | 156.0 | 157.4 |
| C-4b | 152.53 | 152.63 | 151.7 | 151.6 | 151.2 |
| C-5 | 103.57 | 103.52 | 102.7 | 102.6 | 103.0 |
| C-6 | 155.34 | 155.60 | 154.6 | 154.5 | 152.6 |
| C-7 | 146.80 | 146.91 | 146.0 | 145.9 | 146.8 |
| C-8 | 106.53 | 106.43 | 104.8 | 105.2 | 104.3 |
| C-8a | 112.44 | 109.97 | 111.4 | 111.3 | 113.2 |
| C-8b | 102.09 | 102.20 | 101.3 | 101.0 | 101.8 |
| C-9 | 179.56 | 179.61 | 178.9 | 178.6 | 178.8 |
| MeO | 56.91 | 56.89 | 55.2 | 55.8 | 55.8 |
| Glc-1 | 72.32 | 74.04 | 73.0 | 71.2 | 97.4 |
| Glc-2 | 75.63 | 71.32 | 70.1 | 74.3 | 75.2 |
| Glc-3 | 79.89 | 79.60 | 78.9 | 79.0 | 77.0 |
| Glc-4 | 71.53 | 71.39 | 70.6 | 70.6 | 69.6 |
| Glc-5 | 81.66 | 80.55 | 79.8 | 80.9 | 77.6 |
| Glc-6 | 62.15 | 68.64 | 68.3 | 61.2 | 60.5 |
| Api-1 | 109.71 | 109.97 | 109.0 | 108.7 | |
| Api-2 | 77.01 | 76.70 | 75.6 | 75.8 | |
| Api-3 | 79.44 | 79.26 | 78.7 | 79.0 | |
| Api-4 | 74.10 | 74.10 | 73.0 | 73.2 | |
| Api-5 | 65.22 | 64.50 | 63.0 | 73.2 | |
| Rha-1 | 99.9 | ||||
| Rha-2 | 70.3 | ||||
| Rha-3 | 70.5 | ||||
| Rha-4 | 71.8 | ||||
| Rha-5 | 68.4 | ||||
| Rha-6 | 18.1 | ||||
a) Recorded in DMSO-d6 at 201 MHz. b) Recorded in DMSO-d6 at 100 MHz. c) Recorded in DMSO-d6 at 75 MHz.
As described previously, we confirmed the structure of PGX III standard samples currently provided in the market using NMR and MS. It is clear that some of the standard samples distributed as PGX III so far were actually PGX XI. Although PGX III is used to evaluate the quality of Onji by HPLC quantification both in China7) and Japan,8,9) we found the possibility that authentic samples could sometimes be imprecise. In this study we propose using the LC-MS/MS detection method, which discriminates PGX III and XI. As a better way for quality control, we think that both compounds could be used as marker constituents of herbal medicines containing Onji, but further investigation of Onji samples is required.
General procedures UHPLC-MS: UltiMate 3000 RS LC system and Q Exactive Quadrupole-Orbitrap Hybrid Mass Spectrometer (Thermo Fisher Scientific). Column: ACQUITY ultra performance liquid chromatography (UPLC) HSS T3 column (100 × 2.1 mm, 1.8 mm; Waters). Mobile phase: A = 0.1% aqueous formic acid; B = 0.1% formic acid in MeCN. Gradient: 18%B (0–30 min hold), to 95%B (30–30.1, 15 min hold). Flow 0.4 mL/min, column temp 40°C
NMR: JNM-ECZ800 (JEOL) with CH-UltraCOOL™ probe; Temperature: 100°C; 1H-NMR spectra (800 MHz): number of data points, 65536; spectral width, −5 to 15 ppm; acquisition time, 3.27 s; delay time, 2 s; and number of scans, 16. 13C-NMR spectra (201 MHz): number of data points, 65536; spectral width, −25 to 225 ppm; acquisition time, 1.03 s; delay time, 2 s; and number of scans, 2048.
Samples designated as PGX III were purchased from FortopChem Technology (Lot No. FTC170032), AdooQ Bioscience (Lot No. L14750B002) and ChemFaces (Lot No. CFS201701). Onji crude drug (JP17 grade) was purchased from Uchida Wakanyaku Ltd. (Tokyo, Japan).
PGX III1H-NMR (800 MHz, dimethyl sulfoxide (DMSO)-d6) δ: 13.58 (s, 1H, –OH), 7.46 (s, 1H, H-8), 6.85 (s, 1H, H-5), 6.35 (s, 1H, H-4), 4.79 (d, J = 1.9 Hz, 1H, Api-1), 4.65 (d, J = 9.8 Hz, 1H, Glc-1), 3.97 (dd, J = 9.8, 8.9 Hz, 1H, Glc-2), 3.87 (s, 3H, –OMe), 3.85 (dd, J = 11.2, 1.9 Hz, 1H, Glc-6), 3.83 (d, J = 9.3 Hz, 1H, Api-4b), 3.75 (d, J = 2.9 Hz, 1H, Api-2), 3.57 (d, J = 9.4 Hz, 1H, Api-4a), 3.47 (dd, J = 11.2, 6.2 Hz, 1H, Glc-6), 3.37–3.32 (1H, m, Glc-5), 3.36 (d, J = 11.1 Hz, 1H, Api-5), 3.33 (d, J = 11.1 Hz, 1H, Api-5), 3.25 (t, J = 8.9 Hz, 1H, Glc-3), 3.19 (t, J = 8.9 Hz, 1H, Glc-4). 13C-NMR (201 MHz, DMSO-d6) δ: 179.61 (C-9), 164.45 (C-3), 162.27 (C-1), 157.00 (C-4a), 155.60 (C-6), 152.63 (C-4b), 146.91 (C-7), 109.97 (Api-1), 108.28 (C-2), 106.43 (C-8), 103.52 (C-5), 102.20 (C-8b), 94.41 (C-4), 80.55 (Glc-5), 79.60 (Glc-3), 79.26 (Api-3), 76.70 (Api-2), 74.10 (Api-4), 74.04 (Glc-1), 71.39 (Glc-4), 71.32 (Glc-2), 68.64 (Glc-6), 64.50 (Api-5), 56.89 (OMe).
PGX XI1H-NMR (800 MHz, DMSO-d6) δ: 13.54 (s, 1H, –OH), 7.47 (s, 1H, H-8), 6.86 (s, 1H, H-5), 6.34 (s, 1H, H-4), 5.16 (d, J = 1.5 Hz, 1H, Api-1), 4.70 (d, J = 9.8 Hz, 1H, Glc-1), 4.19 (br, 1H, Glc-2), 3.87 (s, 3H, OMe), 3.68 (dd, J = 11.8, 2.5 Hz, 1H, Glc-6), 3.58 (br s, 1H, Api-2), 3.48 (dd, J = 11.8, 5.5 Hz, 1H, Glc-6), 3.42 (t, J = 9.0 Hz, 1H, Glc-3), 3.24 (t, J = 9.0 Hz, 1H, Glc-4), 3.20 (ddd, J = 9.0, 5.5, 2.5 Hz, 1H, Glc-5), 3.15 (d, J = 11.2 Hz, 1H, Api-5), 3.09 (d, J = 9.2 Hz, 1H, Api-4b), 3.01 (d, J = 11.2 Hz, 1H, Api-5), 2.74 (d, J = 9.2 Hz, 1H, Api-4a). 13C-NMR (201 MHz, DMSO-d6) δ: 179.56 (C-9), 164.42 (C-3), 162.18 (C-1), 157.05 (C-4a), 155.34 (C-6), 152.53 (C-4b), 146.80 (C-7), 112.44 (C-8a), 109.71 (Api-1), 108.30 (C-2), 106.53 (C-8), 103.57 (C-5), 102.09 (C-8b), 94.23 (C-4), 81.66 (Glc-5), 79.89 (Glc-3), 79.44 (Api-3), 77.01 (Api-2), 75.63 (Glc-2), 74.10 (Api-4), 72.32 (Glc-1), 71.53 (Glc-4), 65.22 (Api-5), 62.15 (Glc-6), 56.91 (OMe).
This research was supported by AMED under Grant Number 19mk0101102j0102.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.