2014 年 20 巻 1 号 p. 71-77
2014 年 20 巻 1 号 p. 71-77
Loop-mediated isothermal amplification (LAMP), a novel DNA amplification technique, is widely used in molecular diagnosis although it is very easy to be contaminated by the huge amount of its own amplicons. An improved visual LAMP method for the event specific identification of GM rice TT51-1 and a rice endogenous reference gene (sucrose phosphate synthase, SPS) was established. Pre-loaded calcein and manganous ion into the LAMP reaction offered not only an easy way to read results by colour change from orange to green but also a closed- tube system to reduce contaminations caused by amplification products. The LAMP assays could be finished within 60 min, and the limits of detection (LODs) were estimated as low as 10 and 20 copies of rice haploid genomic DNA, respectively, about 10-fold more sensitive than that of conventional PCR. The specificities of the assays were also well evaluated with optimized system and conditions. The developed one-step visual LAMP assays in this study provided a promising tool for the convenient and effective detection of GM crops.
Genetically Modified (GM) crops have greatly changed the current food production and agricultural system in recent years. With the sustained and rapid growth, the global area of GM crops had increased more than 160 million hectares in 2011 (James, 2011). Despite this high utilization rate and promising trend, there are always concerns about the impact of genetically modified organisms (GMOs) on the environment and food safety. To date, more than 50 countries and areas had established separate legislations and regulations for labeling the presence of GM ingredients (Gru—re & Rao, 2007). Although the explosion of GM crop yields and relevant labeling policies significantly increased the demand for rapid and accurate testing procedures, the contradiction between complicated, time-consuming testing procedures and the growing of the number and types of GMOs is increasingly outstanding. Therefore, developing quick, accurate, high sensitive and efficient detection methods for GMOs has turned to be more important in the near future.
Rice (Oryza sativa), as staple food in many Asian countries, is the main dietary source for more than half the world's population. So far, several GM rice events had been developed and approved for commercialization with various agronomic traits, such as insect resistance, bacterial resistance and herbicide tolerance (Bajaj & Mohanty, 2005). TT51-1 (synonym BT63), an insect-resistant transgenic variety, is the first GM rice event granted a safety certificate in China, which was created though microprojectile bombardment strategy based on the elite Chinese cytoplasmic male sterile (CMS) restorer line Minghui 63 (Lu, 2010). Additional five GM rice lines were used in this study for specific evaluation, including KeFeng6, IIKF6, KMD1, M12, and Wan21B. KeFeng6, IIKF6 and KMD1 lines were integrated with cry genes, while M12 and Wan21B lines carrying the Xa21 gene were highly resistant to rice bacterial blight (RBB). More recently, the integrated elements and boundary regions of TT51-1 were isolated and characterized, which facilitated the establishment of event-specific testing method and reference plasmid construction (Cao et al., 2011).
Polymerase Chain Reaction (PCR)is the main approach for GMO detection due to its superiorities in sensitivity, efficiency and applicability compared to protein-based methods (Holst-Jensen et al., 2003; Marmiroli et al., 2008). Although several alternative amplification methods which can geometrically amplify targets through different mechanisms had also been developed for molecular identification, for instance, self-sustained sequence replication (Guatelli et al., 1990), nucleic acid sequence-based amplification (NASBA) (Compton, 1991; Morisset et al., 2008), strand displacement amplification (SDA) (Walker et al., 1992), and rolling cycle amplification (RCA) (Lizardi et al., 1998), they did not achieve significant improvement in sensitivity, specificity and facilitation to become replacement methods for routine analysis of GMO. Additionally, many improved technologies that can provide closed-tube solutions were also reported, which using special fluorescent-labeled probes to combine the amplified end products, but these methods did not have advantages in terms of convenience and low-cost (Holland et al., 1991; Zhang et al., 1998; Whitcombe et al., 1999).
Loop-mediated isothermal amplification (LAMP) is a novel DNA amplification technique originally developed by Notomi et al. which utilize the strand displacement activity of Bst DNA ploymerase and a set of specially designed primers to generate tremendous amount of hairpin structures in various lengths (Notomi et al., 2000). The common primer set is composed of two pairs of primers that recognize six regions of the target sequence (Fig. 1A). And the reaction can be accelerated by the application of extra loop primers that hybridizing to the stem-loops (Nagamine et al., 2002). Comparing to PCR method, LAMP reaction is performed at a constant temperature (60 ∼ 65°C) which is more suitable for in-field and rapid testing with simple isothermal instruments. Moreover, the LAMP results could be directly judged with naked eyes by turbidity or addition of fluorescent dyes instead of agarose gel electrophoresis analysis (Mori et al., 2001; Guan et al., 2010). Due to the higher specificity, sensitivity and amplification efficiency, LAMP had been widely used for the diagnosis of viruses (Bista et al., 2007; Yoneyama et al., 2007) and pathogenic microorganisms (Iwamoto et al., 2003; Kuboki et al., 2003; Hara-Kudo et al., 2008), but it can hardly be seen on the application for GM crops up to now (Fukuta et al., 2004; Lee et al., 2009; Liu et al., 2009; Guan et al., 2010; Chen et al., 2011).

Primer design for LAMP assays. Panels A showed the schematic diagram of LAMP primer design, where c represents a complementary sequence. Panels B and C demonstrated the primer locations of SPS and TT51-1 assays. Nucleotide sequences were indicated by lines. Inner primers (FIP and BIP), outer primers (F3 and B3) and loop primers (LF and LB) were used in this study.
However, the main weakness of conventional LAMP is that the workplace is easy to be contaminated by the extremely large amount of amplicons. Thus, a closed-tube system is essential for LAMP reaction to minimize the potential cross-contaminations and reduce false positive results. Recently, an improved LAMP strategy had been reported which applied the manganous ion and calcein, a fluorescent metal indicator, into the reaction system so that the result could be readily distinguished by colour change from orange to green without opening tube after amplification (Tomita et al., 2008). And this novel visual LAMP method had been successfully applied to Mycobacterium (Zhu et al., 2009) and Brucella (Pan et al., 2011). In this paper, we optimized this type visual LAMP assay with one-step operation and evaluated the real-time turbidity, specificity and sensitivity of event-specific detection of GM rice TT51-1. These results indicated that this one-step visual LAMP is an easy and reliable solution for molecular screening for GM ingredients.
Plant Materials The seeds of GM rice events (TT51-1, KeFeng6, KMD1, M12, Wan21B, and IIKF6) were provided by the Qinhuangdao Entry-Exit Inspection and Quarantine Bureau (Qinhuangdao, China). Non-GM seeds of rice, soybean, maize, rape, and wheat were obtained from local market in Tianjin, China.
DNA Extraction Genomic DNA were isolated and purified using the GMO DNA extraction kit (Tiangen Biotech Co. Lt., Beijing, China) according to the manufacturer's instructions. DNA purity and concentration were measured by the NanoDrop 1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). And the quality was evaluated by 1% (w/v) agarose gel electrophoresis.
Primer Design The online software Primer Explorer V4 (http://primerexplorer.jp/elamp4.0.0/index.html) was employed for LAMP primer designing. Sucrose phosphate synthase (SPS) gene (Genebank Acc. No. U33175) was designated as the rice endogenous reference gene in this study (Fig. 1B). The event-specific LAMP primers of TT51-1 were designed based on the downstream junction of the integrated sequence (Genebank Acc. No. EU880444), including three pairs of primers (F3, B3, FIP, BIP, LF and LB) (Fig. 1C). To compare the sensitivity of LAMP to that of conventional PCR, the primers of routine qualitative PCR testing were selected for SPS (SPS-F/R) and event specific detection of TT51-1 (TT51-1-F/R) (MA of PRC, 2009). All of the primers were synthesized by Invitrogen (Invitrogen Co. Ltd., Shanghai, China) and listed in Table 1.
| Primer name | Sequences (5′-3′) | Length | |
|---|---|---|---|
| LAMP | SPS-F3 | GATGCGTTTGTCCCGTCC | 18 |
| SPS-B3 | GTAACCACCCAATCCTCCTC | 20 | |
| SPS-FIP | AGCTCGACCACCACCACT-TGCAGTGGCAGTGCAAGCAT | 38 | |
| SPS-BIP | GGCCGCTCTTGTCTGCACAG-CCCCATGGCAACAACACG | 38 | |
| SPS-LF | GCGCGCGTCGATCGATAA | 18 | |
| TT51-1-F3 | AATGACCGCTGTTATGCG | 18 | |
| TT51-1-B3 | TTTTGGAACATATGAGTGGTAG | 22 | |
| TT51-1-FIP | CCTCTAGAGTCGACCTGCAG-CCATTGATTTGTAGAGAGAGAC | 42 | |
| TT51-1-BIP | CGAGTGCTGGGGCAGATAAG-CGTCCAGAAGGAAAAGGAAT | 40 | |
| TT51-1-LF | CATGCCCGCTGAAATCACC | 19 | |
| TT51-1-LB | GTAGTGGTGGGGCTACGAAC | 20 | |
| Conventional PCR | SPS-F | TTGCGCCTGAACGGATAT | 18 |
| SPS-R | GGAGAAGCACTGGACGAGG | 19 | |
| TT51-1-F | AGCAGAACTTTAACCCCCGAA | 21 | |
| TT51-1-R | AGAGCCTCGTTGGATTTCTTACAT | 24 |
LAMP Assay LAMP was carried out in a 25 µl volume mixture containing 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2 SO4 , 0.1% Triton X-100, 6 mM MgSO4 , 0.8 mM each dNTP, 2.5 µg BSA (Promega Corporation, Madison, USA), 50 µM Calcein, 0.5 mM MnCl2, 40 pmol each of inner primers, 10 pmol each of outer and loop primers, 8 units of the Bst DNA polymerase large fragment (New England Biolabs, Ipswich, Beverly, MA), and 10 ng template DNA. The TT51-1 assay was incubated at 61°C, while the optimum temperature of SPS is 64°C. All LAMP reactions were sustained for 70 min and terminated by heating at 80°C for 3 min.
PCR Reaction A total volume of 25 µL PCR system comprised 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.25 mM each dNTP, 0.1 µM each primer, 1 unit Taq polymerase (TaKaRa biotechnology Co., Ltd, Dalian, China) and 10 ng template DNA. The PCR reactions were performed on a Veriti 96-Well thermal cycler (Applied Biosystems, Foster City, CA, USA) according to the following program: 94°C for 2 min, 35 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 7 min. PCR products of SPS (277 bp) and TT51-1 (274 bp) were analyzed by 2% (w/v) agarose gel electrophoresis with Gel-Red staining.
Detection of LAMP Products The real-time turbidity of the LAMP reaction was detected by a Loopamp real-time turbidimeter LA-320 C (Teramecs, Kyoto, Japan). To determine the efficiency and optimal temperatures, the LAMP assays were incubated for 90 min at 60 to 65°C. The positive results of LAMP could be directly determined with the naked eye in which colour would turn green instead of the original orange. Positive tubes could also be effectively distinguished under ultraviolet (UV-366 nm) which emitting strong fluorescence. Agarose gel electrophoresis is the necessary approach for the confirmation of characteristic ladder-like bands. Several GM rice events and non-transgenic crops were used for the specificity evaluation. And the sensitivity of the one-step visual LAMP was surveyed by amplifying serially diluted TT51-1 genomic DNA samples.
Primer design and optimization of LAMP Available options of primer design for TT51-1 were very limited due to the needs of event-specific detection. Here, we utilized the loop primers which could greatly enhance the efficiency of LAMP and reduce reaction time, although four primers (two inner and two outer) are capable of LAMP amplification in general (data not shown). LAMP primers of the rice endogenous reference gene were located on the SPS gene, which occupied a 206 bp target sequence. Considering of the practicability of LAMP, non-denatured DNA samples were selected as templates in this work. The optimal reaction temperature was determined by comparing the gradient temperatures ranging from 60°C to 65°C, and the results corresponded to 61°C and 64°C for TT51-1 and SPS LAMP assays, respectively. The concentration of magnesium was increased to 6 mM and bovine serum albumin (BSA) was added in each reaction. Also, the ratios between inner (FIP and BIP), outer (F3 and B3) and loop primers were also optimized. The proportion of calcein and manganous ion were finally determined as 50 µM and 0.5 mM in each reaction.
Real-time turbidity of LAMP In order to identify the threshold time of the LAMP amplifications, TT51-1 and SPS primer sets were tested by real-time turbidity detection with 10 ng TT51-1 genomic DNA template. As shown in Fig. 2A and 2B, the threshold of SPS assay occurred at 36 min, while the TT51-1 event specific LAMP was at 60 min. The monitoring results could also be simply visualized by naked eye under daylight or UV (Fig. 2C and 2D). The typical characteristic ladder-like bands was confirmed by agarose gels electrophoresis (Fig. 2E). These results suggested that the efficiency of developed assays were sufficient for rapid testing of target DNAs.
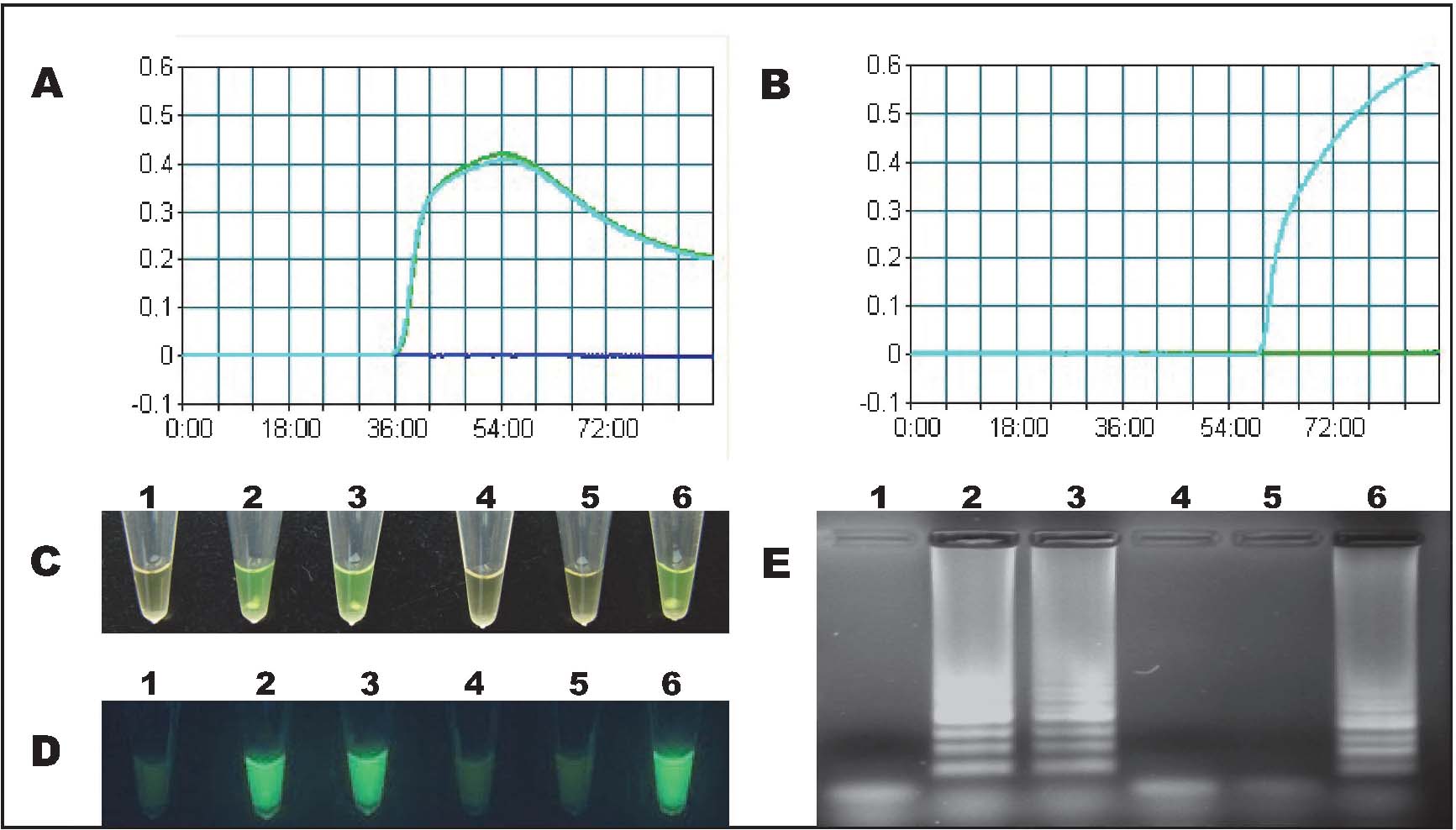
Real-time detection of LAMP assays. Panels A and B showed the results of real-time turbidity detection of SPS and TT51-1, respectively. Template order: blue curve, NC; green curve, non-GM rice; cyan curve, TT51-1. Panels C, D and E showed the results of LAMP in the form of normal, under UV (366 nm) and 2% agarose gel electrophoresis. Lanes 1, 2, and 3, SPS; lanes 4, 5, and 6, TT51-1; Template order: lanes 1 and 4, NC; lanes 2 and 5, non-GM rice; lanes 3 and 6, TT51-1.
Specificity of LAMP Six GM rice events (TT51-1, KeFeng6, KMD1, M12, Wan21B, and IIKF6) and five non-GM plants (rice, maize, soybean, rape, and wheat) were used to evaluate the specificities of the established assays. Ten nanogram of genomic DNA was employed for single LAMP reaction. As a result, the primer set of SPS successfully identified the non-transgenic rice and all GM rice events while no positive reaction was found from other non-GM plants and negative control (NC). In the TT51-1 assay, the characteristic ladder-like products and colour change were observed only when the template is TT51-1 genomic DNA. These results were displayed in form of normal, under UV light, and after agarose gel electrophoresis (Fig. 3). It worth noting that the high consistency between colour change and gel electrophoresis analysis confirmed that visual detection is a credible and exact way to read results.

Specificity test of LAMP assays. Panels A, B, and C showed the results of SPS, and panels D, E, and F showed the results of TT51-1 in the form of normal, under UV (366 nm) and 2% agarose gel electrophoresis, respectively. Lane 1, NC; lanes 2 - 6, non-GM rice, non-GM maize, non-GM soybean, non-GM rape, and non-GM wheat; lanes 7 - 12, TT51-1, KeFeng6, KMD1, M12, Wan21B, and II KF6.
Sensitivity of LAMP High sensitivity is a requirement for practical GMO detection due to the poor quantity DNA derived from processed food with GMOs. To assess the limits of detection (LODs) of LAMP, TT51-1 genomic DNA was serially diluted ranging from 20000, 2000, 200, 100, 20, 10, and two copies haploid genome per reaction tube. Replicate experiments revealed that the LODs of SPS and TT51-1 were estimated at 10 and 20 copies, respectively, which was approximately 10-fold more sensitive than that of conventional PCR method. Results with colour change and gel electrophoresis analysis can be seen in Fig. 4. The relatively low LOD was attributed to the design of the LAMP primers and the structural of the target DNA sequence. All of above results indicated that, to a certain extent, the developed LAMP assays have good potential to replace the GMO routine testing methods in both laboratory and field applications.
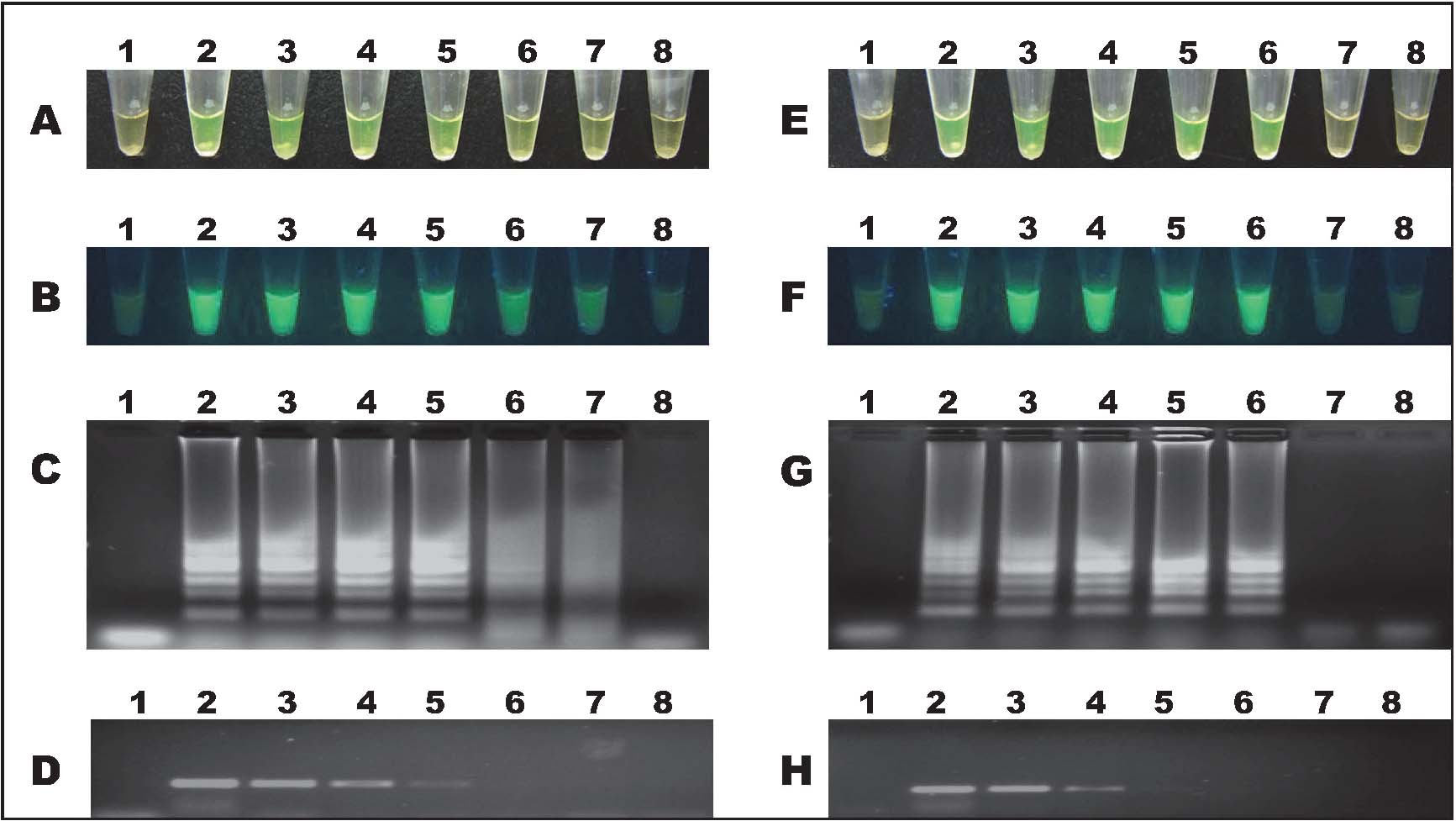
Sensitivity test of LAMP assays. Panels A, B, and C showed the results of SPS, and panels E, F, and G showed the results of TT51-1 in the form of normal, under UV (366 nm) and 2% agarose gel electrophoresis, respectively. Panel D and H were the agarose gel electrophoresis results of SPS and TT51-1 conventional PCR. Lane 1, NC; lanes 2 - 8, 20000, 2000, 200, 100, 20, 10, and two copies haploid genomic DNA, respectively.
In this study, we developed the one-step visual LAMP method for the detection of GM rice TT51-1 and endogenous reference gene SPS. Evaluation of the real-time turbidity, LODs and specificities of these methods revealed that the developed LAMP assays are rapid, sensitive, and reliable approaches for GM rice TT51-1 detection. As expect, the results could be easily distinguished by colour change from orange to green. Moreover, the most prominent characteristic of the one-step visual LAMP assay is that it offered a closed-tube system instead of opening tube after amplifications which could minimize the risk of workplace contamination.
Notably, there was similar work has been published during this manuscript submission (Chen et al., 2012). Although they also reported the TT51-1 specific detection method based on the LAMP strategy, there were still important distinctions between our work and that paper. First and most important was that this study provides a closed-tube system to eliminate contaminations caused by end products. Given that a LAMP reaction could generate huge amount of amplicons, opening the finished tubes is the major reason for false positive results especially when using SYBR green in LAMP experiments, because opening tube to add fluorescence dye after reaction is unavoidable. Secondly, the primer set for TT51-1 detection is dissimilar, where the utilization of loop primers made it more effective to amplify targets. Thirdly, the components and optimal conditions of the LAMP assays are quite different. All the LAMP reactions in this study were carried out with undenatured template DNA to pursue a convenient way for practical applications. Last but not least, the SPS gene was designated as endogenous reference gene instead of PLD gene. Taken together, we have reason to believe that the developed LAMP assays in this work are suitable for GM rice detection with the advantages of visual, simplicity, rapidity and better sensitivity.
Data from this paper proved that the developed one-step visual loop-mediated isothermal amplification (LAMP) assay is a promising tool for convenient and effective detection of genetically modified rice event TT51-1. Besides high sensitivity and specificity, the most important improvement is that it offered a closed-tube system instead of opening tube after amplifications, which would greatly reduce the workplace contamination.
Acknowledgment This work was supported by Agricultural Science and Technology Achievements Transformation Funds Project of China (No. 2011GB2A100011), President funding of Tianjin Academy of Agricultural Sciences (No. 13002) and National Natural Science Foundation of China (No. 31000356).