2014 Volume 20 Issue 4 Pages 899-904
2014 Volume 20 Issue 4 Pages 899-904
The spoilage and pathogenic bacteria in the sporadic microbial spoilage of Sichuan Pickle were investigated by 16S rRNA libraries. The pathogenic Vibrio penaeicida (39.19%), the spoilage and pathogenic Pseudomonas fluorescens (6.76%) were detected in initial spoilage stage. While, in middle spoilage stage they shifted to the pathogenic V. penaeicida (51.25%), the spoilage and pathogenic Pse. fluorescens (3.75%), the spoilage Pse. Chlororaphis (7.5%) and then to the pathogenic V. penaeicida (30.59%), Halomonas variabilis (10.59%) and Arcobacter marinus (5.88%), the spoilage Lactobacillus alimentariu (5.88%) in latter spoilage stage. The undesired Pseudoalteromonas nigrifaciens, Psychrobacter alimentarius, Marinomonas, Cobetia marina, Celerinatantimonas and V. litoralis were also detected in the spoilage process. When compared at species level, the community similarity coefficients of three samples were from 0.6122 to 0.5165 and ascended to 0.2359. These results are very useful to design the effective strategies to control or eliminate the microbial spoilage of Sichuan Pickle.
Sichuan Pickle, mainly produced in home or small scale with pottery in Sichuan of China, is a typical representative of Chinese traditional vegetable fermentation art. Its fermentation is a natural process in which the spontaneous fermentation results from the competitive activities of about 10−6 – 10−7 CFU/mL indigenous lactic acid bacteria in old salt brine together with a variety of microbes presented on the raw vegetable. In many regions of China, Sichuan Pickle is normally served as a key flavoring for Sichuan cuisine or used as an appetizer because of its unique flavor. Unlike sauerkraut and kimchi which use direct salting to withdraw juice from the cabbage, Sichuan Pickle is a type of brinesalted and lactic acid spontaneously fermented vegetable product. The vegetables, such as leaf mustard, cabbage, radish, bamboo shoot, tender ginger and chili, are immersed into the about pH 4.5 old rine with 6% – 8% (w/v) salt and then are spontaneously fermented (Xiong et al., 2012). In recent years, the Sichuan Pickle manufacturing process has carried out on industrial scale by some enterprises from Chengdu plains of Sichuan, China. Unfortunately, these producers often meet sporadic microbial spoilage of pickle in top layer of pickle pool.
Microbial spoilage of food is a complex event, in which the combination of microbial and biochemical activities may interact, and can be considered as any change which renders a product unacceptable for human consumption (Huisin 't Veld et al., 1996). In general, Sichuan Pickle fermentation on industrial scale is also a natural process. Its spontaneous fermentation is only mediated by the competitive activities of the microbes loaded on the vegetable and from the wall of pickle pool, plastic film covering and pickle production environment, but without the lactic acid bacteria from the old salt brine. Therefore, the fermentation process is easy to be disturbed by some spoilage and pathogenic microbes, specially the top layer which usually exhibits the same spoilage phenomenon. To minimize the top layer pickle spoilage, manufacturers of Sichuan Pickle have aspired to implement strategies to control or eliminate the microbes responsible for spoilage. Regretfully, their efforts were fruitless because the microbes implicated in Sichuan Pickle spoilage were unknown until the present.
In the spontaneous fermentation ecosystem, whether or not the spoilage microbial groups will contribute to spoilage mainly depends on their living conditions and their competitions. And their community succession associated with the fermentation process was reported to significantly affect the type of spoilage (Doulgeraki et al., 2012). In fact, several studies have focused on the microbial diversity and community succession in the industrial scale spontaneous fermentation process of Sichuan Pickle (Xiong et al., 2012). However, the spoilage-related microbes involved in the fermentation process of sporadic top layer spoilage pickle are not yet investigated. Therefore, the aim of current work was to mainly investigate the spoilage and pathogenic bacteria associated with the sporadic top layer spoilage process during the spontaneous fermentation of Sichuan Pickle by 16S rRNA gene libraries. The information gathered may be useful to design the effective strategies to control or eliminate the microbial spoilage of Sichuan Pickle during the industrial fermentation.
Spoilage Sichuan Pickle sampling The experiments was carried out on industrial scale at the Zhuongleeji Food Co. Ltd (Sichuan, China). The abnormal fermentation Sichuan Pickle in the top layer of fermentation pool was selected to investigate the spoilage and pathogenic bacteria. The salt brine fermented for 7 days (SP07; pH6.3, 20°Cand 5% (w/v) NaCl), 22 days (SP22; pH5.8, 20°C and 4.8% (w/v) NaCl) and 60 days (SP60; pH5.0, 20°C and 4.6% (w/v) NaCl) were respectively collected in the sterile tubes from the different pickle pool and then analyzed after they were mixed.
DNA extraction and construction of 16S rRNA library The microbes in the 50 mL salt brine were collected by centrifugation at 8000 revolutions per minute for 10 min at 4°C. And the resulting pellet was re-suspended in 30 mL PBS buffer (pH 7.4) and vortexed for 5 min. The suspension was centrifuged again at the above condition, and then the pellet was re-suspended in 0.5 mL PBS buffer and transferred to a 1.5 mL tube. The microbial cell disruption and extraction of total DNAs were performed by the methods described as Marshall (Marshall et al., 2008). The construction of 16S rRNA library was as previously described by Xiang (Xiang et al., 2013).
Taxonomic assignment and phylogeny The taxonomic assignment was given by RDP classifier program with a bootstrap score of 80%. The nearly full-length 16S rRNA without chimeras were compared against those deposited in GenBank (www.ncbi.nlm.nih.gov/BLAST/) to identify their closest phylogenetic relatives. Phylogenetic analysis based on the 16S rRNAs was constructed with MEGA 5.0 by the Neighbor-Joining method with the Kimura two-parameter model (Tamura et al., 2011).
Statistical analysis BioEdit software was employed to carry out the paired comparison of all 16S rRNA sequences, and the sequences, indicating at least 97% similarity, were grouped into the operational taxonomic units (OTUs), while any remaining sequences were grouped alone. The rarefaction analysis was performed by the Analytic Rarefaction 1.3 calculator based on the numbers of OTUs versus the numbers of clones (Sundarakrishnan et al., 2012). The Sobs, Schao1, SACE, Shannon diversity (H), Simpson and Coverage were calculated by Estimates Win 8.20 software. The Sobs is the number of observed OTUs. The SACE provides an estimate based on the relative number of OTU that occur only once, no more than 10 times, and more than 10 times. The SChao1 takes into account the relative number of three classes: single OTU, those that appear twice, and the total number. The Simpson index measures the distribution of clones among OTUs. The H index measures the probability of correctly guessing the OTU identity of a randomly selected clone. The Coverage is the ratio of the OTUs represented and the OTUs inhabited in the samples. Microbial community relationship of three samples was expressed with Baroni-Urbani and Buser similarity index estimated by DPS 7.5.
Nucleotide sequence accession numbers The represent 16S rRNA sequences of every OTU were deposited in the GenBank at the NCBI. The GenBank ID: KF601962 - KF601977.
16S rRNA libraries of spoilage Sichuan Pickle Each sample was used to produce a 16S rRNA library. A total of 239 bacterial clones were recovered from three cover brines, 74 clones in SP07 sample, 80 clones in SP22 sample and 85 clones in SP60 sample. The 16S rRNA sequences that showed 97% or more similarity were sorted into OTUs. It was determined that there were 7 OTUs in SP07, 6 OTUs in SP22 and 7 OTUs in SP60, and that all OTUs contained multiple clones.
Spoilage and pathogenic bacteria diversity The richness of the spoilage and pathogenic bacteria presented in the samples was estimated by rarefaction curves. The curves were respectively obtained by plotting the number of OTUs against the number of clones (Fig. 1). The obvious decrease in the rate of OTUs detection indicated that the bacterial richness of three brine samples was almost completely revealed, and was very close to the true bacterial diversity. This conclusion was further verified by calculating 100% coverage of tested samples (Table 1). Biodiversity was calculated for bacterial populations using Shannon-wiener (H) and Simpson dominancy index. The H index and the Simpson index respectively varied from 1.25 to 1.69 and from 2.47 to 4.91 for three libraries (Table 1). Maximum OTU richness was respectively estimated with SChao1 and SACE, and it revealed that SChao1 7 and SACE 7 for SP07, SChao1 6 and SACE 6 for SP22, SChao1 7 and SACE 7 for SP60 (Table 1). Based on these estimations, almost 100% of the spoilage and pathogenic bacterial diversities were covered by the applied sampling survey.
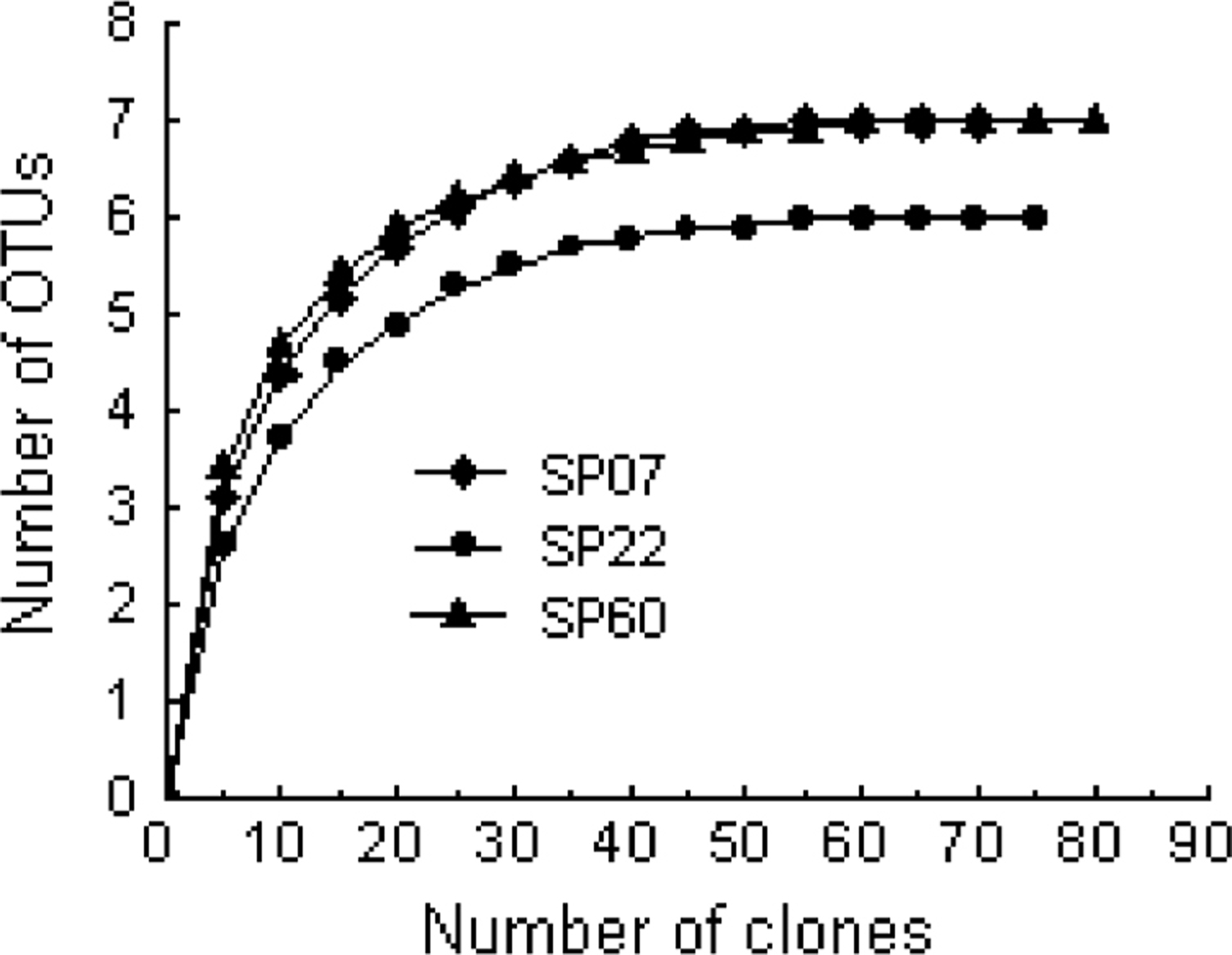
Rarefaction analysis of spoilage and pathogenic bacteria in SP07, SP22 and SP60 sample based on the 16S rRNA libraries, displaying the number of OTUs detected versus the number of sequences analyzed.
| Sample | Clones | Sobsa | Schao1 | SACE | Coverage | H | Simpson |
|---|---|---|---|---|---|---|---|
| SP07 | 74 | 7 | 7 | 7 | 100 | 1.55 | 3.68 |
| SP22 | 80 | 6 | 6 | 6 | 100 | 1.25 | 2.47 |
| SP60 | 85 | 7 | 7 | 7 | 100 | 1.69 | 4.91 |
Spoilage and pathogenic bacteria distribution and phylogeny All 16S rRNAs without chimeras were accurately assigned to genus or species level by RDP MultiClassifier. In SP07sample, seventy four 16S rRNA sequences respectively were fell into the genera Vibrio, Pseudoalteromonas, Oceanobacillus, Marinomonas, Psychrobacter, Pseudomonas and Leuconostoc. V. penaeicida (39.19%), Psa. nigrifaciens (20.27%) and O. sojae (13.51%) were predominant in the spoilage initial process. Eighty 16S rRNAs retrieved from the SP22 were respectively classified to the Vibrio, Cobetia, Pseudomonas, Oceanobacillus and Lactobacillus. And V. penaeicida (51.25%) and C. marina (12.5%) were dominant. Eighty five 16S rRNAs in SP60 were respectively identified to the Vibrio, Cobetia, Halomonas, Lactobacillus, Arcobacter, Oceanobacillus and Celerinatantimonas. V. penaeicida (30.59%), C. marina (24.71%), H. variabilis (10.59%) and Lac. sakei (14.12%) were prevalence in the spoilage latter process (Fig. 2).

The distribution of spoilage and pathogenic bacteria in SP07, SP22 and SP60 sample.
Molecular phylogeny increasingly supports our understanding to the microbial relationships in environment or fermentation sample (Xiang et al., 2013). In current work, all 16S rRNA sequences were submitted to NCBI public database to infer a possible phylogenetic classification. The BLAST searches revealed that most sequences shared more than 98 % similarity with strains in public database at the 16S rRNA level. Eichler demonstrated that the microorganisms with a similarity higher than 98% of 16S rRNA sequences could be clustered into same species (Eicher et al., 2006). While, the bacteria represented by clone SP07 - 34, SP07 - 46, SP60 - 33 and SP60 - 66 showed 97% similarity with type strains V. penaeicida DSM14398T, Leu. gelidum DSM 5578T, H. variabilis DSM 3051T and C. diazotrophica JG2 - 2T respectively. To further disclose the microbial taxonomic position and relationship, the phylogenetic tree based on the 16S rRNA representative sequences was constructed by neighbor-joining method (Fig. 3). The tree revealed the phylogenetic affiliation of spoilage and pathogenic bacteria associated to the spoilage process of Sichuan Pickle in the top layer of pickle pool during the spontaneous fermentation.

The phylogenetic tree based on the complete16S rRNA sequences of spoilage and pathogenic bacteria in SP07, SP22 and SP60 sample by using the neighbor-joining method. The scale bar corresponds to 0.02-estimated nucleotide substitution per sequence position. Bootstrap values from 1000 replicates are included.
Spoilage and pathogenic bacterial community succession In general, the microbial succession is a universal phenomenon happened in spontaneous fermentation process. Many studies have examined patterns of microbial succession in the kimchi (Chang et al., 2008), traditional fermented mustard (Chao et al., 2009), Vietnamese alcohol starter (Thanh et al., 2008), and Chinese traditional liquor fermentation process (Xiang et al., 2013). During the spoilage process of Sichuan Pickle in the top layer of pickle pool, the bacterial community similarity coefficient in three samples was from 0.6122 to 0.2359, and followed to 0.5165 when compared at the species level (Table2). This result suggested that the spoilage and pathogenic bacterial community in the tope layer Sichuan Pickle has already shifted because of microbial competition increases, pH decreases, and oxygen decreases associated with fermentation proceeding. Furthermore, this phenomenon was also proclaimed by the variations of H index, simposon index and spoilage microbial distribution in three samples (Table 1 and Fig. 2).
| Similarity coefficient | SP07 | SP22 | SP60 |
|---|---|---|---|
| SP07 | 1.0000 | 0.6122 | 0.2359 |
| SP22 | 1.0000 | 0.5165 | |
| SP60 | 1.0000 |
In the traditional vegetable pickle fermentation, lactic acid bacteria play an important role. Leu. mesenteroides, Leu. citreum, Lac. plantarum, Lac. brevis, Lac. curvatus and Lac. sakei are typical functional bacteria for sauerkraut, Sichuan Pickle and kimchi fermentation, and they are initially present in low numbers and followed by a rapid increase after three days of fermentation (Wouters et al., 2013). Interestingly, in the salt brine of top layer industrial Sichuan Pickle spoilage process, they were not detected or not prevalent in three samples, only Leu. gelidum (4.05%) in SP07, Lac. graminis (6.25%) and Lac. sakei (3.75%) in SP22, and Lac. sakei (14.12%) and Lac. alimentarius (5.88%) in SP60. Leuconostoc dominates the early stage of traditional vegetable fermentation and produces lactic and acetic acid that inhibit harmful organisms (Kim et al., 2000). Although Leu. gelidum could also soon initiate the vegetable fermentation as Leu. Mesenteroides (Kim et al., 2000), Leu. gelidum was not dominant in the early fermentation stage of top industrial Sichuan Pickle. Therefore, it could not produce adequate lactic and acetic acid to inhibit harmful bacteria. Furthermore, the vegetable juice extracted by salt took some part salt adhered on the top layer vegetable surface into the middle layer and the bottom layer brine because the salting vegetable in the top layer usually floated over the salt brine, and resulted in that the salt concentrate (5% v/v) was lower than that in the middle and bottom layer (8% v/v). The low salt made a more favorable environment for development of bacteria. And thus the spoilage and pathogenic bacteria from the vegetable material, wall of pickle pool, plastic film covering and pickle production environment were easy to grow and develop on the top layer industrial Sichuan Pickle. This is main reason why the Sichuan Pickle sporadic spoilage often takes place on the top layer.
The investigation of microbes able to compromise the quality and safety of foods is a major concern for the food industry and the administration (Fernandez-No et al., 2011). In current work, the bacterial community in the top layer industrial Sichuan Pickle were investigated in the different fermentation stage. Surprisingly, some spoilage or pathogenic bacteria were the dominant instead of the expected dominance of the lactic acid bacteria in the fermenting vegetable, and their community structures have had succession associated with the spoilage process. The pathogenic V. penaeicida was from 39.19% in SP07 to 51.25% in SP22 and followed to 30.59% in SP60. The V. penaeicida is one of the most serious pathogens for penaeid shrimp aquaculture (Goarant et al., 2006). Although there is few reports in fermented vegetables, its appearance maybe bring potential harm owing to producing protein exogenous toxin and causing food spoilage (Saulnier et al., 2000). Pse. fluorescens and Pse. chlororaphis are the most frequently found dominant species of many vegetables in the field (Franzetti et al., 2007). They have been known to be responsible for food spoilage, however their distribution in the fermentation food ecosystem remains relatively unknown (Franzetti et al., 2007). Pse. fluorescens is also pathogenic bacterium, and it usually affects patients with the compromised immune systems (Franzetti et al., 2007). In current study, Pse. fluorescens was only found to be present in SP07 (6.76%) and SP22 (3.75%). In food industry, Pse. chlororaphis, 7.5% only in SP22, is considered as a major producer of the volatile compounds responsible for off-flavor compounds, and it plays a primary role in the process of foodstuff spoilage (Franzetti et al., 2007). The pathogenic H. variabilis and A. marinus were only detected in the SP60 and they were 10.59% and 5.88%, respectively. The genus Arcobacter has become increasingly important concern in recent years because its members have been considered emergent enteropathogens and potential zoonotic agents (Collado et al., 2011). H. variabilis could be isolated from the various aquatic and terrestrial euryhaline habitats (Okamoto et al., 2004). Although it is not yet clear whether H. variabilis is pathogenic bacterium or not, many literatures indicate the genus Halomonas is a new potential pathogen to human (Stevens et al., 2013). Lac. alimentariu, 5.88% only in SP60, is also found to be the specific spoilage microbe affected various high-salt food product qualities and it can survive in acidic and high-salt environments (Lyhs et al., 2001). The undesired bacteria Psa. nigrifaciens, Psy. alimentarius, M. arenicola, Cob. marina, Cel. diazotrophica and V. litoralis were also detected in the Sichuan Pickle spoilage process. However, whether they could also cause the food spoilage or foodborne illness or not is unknown so far.
Usually, food spoilage is an interaction process of microbial community succession and their biochemical activities (Huisin't Veld et al., 1996). The current study has provided us a dynamics of spoilage and pathogenic bacteria associated with fermenting in the top layer Sichuan Pickle. Although their biochemical activities responsible for the Sichuan Pickle spoilage should receive further studies, the present information gathered is useful to design the effective management to control or eliminate the spoilage and pathogenic bacteria during the fermentation of Sichuan pickle.
Acknowledgements This work was financially supported by the grant from the Sichuan pickle industry chain project of Sichuan province (2012NZ0002-8), the applied basic research program of the science and technology department of Sichuan, China (2014JY0045) and the key research program of the education department of Sichuan, China (14ZA0110).