2014 年 20 巻 4 号 p. 905-913
2014 年 20 巻 4 号 p. 905-913
A total of 270 food samples were obtained from twelve free markets (149 samples) and seven supermarkets (121 samples) in Thailand from September 2010 to February 2012. The samples were divided as follows: 51 meat, 37 fish/seafood, 38 vegetable, 11 fermented food and 133 tofu samples. The samples were examined for the presence of Salmonella spp.. Salmonella spp. were detected in 28% of the free market samples and in 7% of the supermarket samples. Meat samples had the highest contamination rates: 78% of the free market and 40% of the supermarket samples. Of the fish/seafood, vegetable, fermented food and tofu samples from the free markets, 41%, 6%, 25%, and 7%, respectively, tested positive for Salmonella spp., whereas the corresponding contamination rates were 5%, 5%, 0% and 0% for the supermarket samples. The resistance of the isolates towards sixteen antibiotics was tested. A high percentage of the isolates were resistant to streptomycin, tetracycline or ampicillin: 78%, 59% and 51%, respectively. In total, 152 isolates were resistant to at least one antibiotic, and of these, 98 (64%) were resistant to at least three antibiotics (multidrug-resistant). The most frequently isolated serovars were Corvallis, Rissen and O4: i:-. Random Amplified Polymorphic DNA (RAPD) was used for grouping 149 isolates into five groups. The grouping of the isolates is baseline information to emphasize the usefulness of subtyping Salmonella spp. isolated from several categories of retail foods obtained from free markets and supermarkets by using antibiotic resistance studies and RAPD grouping.
Salmonella enterica, a leading bio-agent of human gastroenteritis in both developed and developing countries, causes millions of human illnesses and significant economic losses worldwide. It has been reported as a contaminant of several foods, including poultry, pork, beef, fish, seafood, egg, milk, cheese, fresh fruits and vegetables (Salleh et al., 2003; Bangtrakulnonth et al., 2004; Shabarinath et al., 2007; Minami et al., 2010; Sant'Ana et al., 2011). The emergence and spread of antibiotic-resistant Salmonella, particularly multidrug-resistant (MDR) strains, is a major public health concern. Most infections with MDR Salmonella are acquired by eating contaminated foods of animal origin (White et al., 2001). Antibiotic resistance of Salmonella spp. has resulted in failed treatments for salmonellosis (Butt et al., 2003). The isolation of antibiotic-resistant Salmonella spp. from chicken meat and human stool in Thailand has been previously reported (Boonmar et al., 1998; Kulwichit et al., 2007).
S. enterica contains more than 2500 serotypes in the Kauffmann-White Schema. Serotyping based on the differentiation of O and H antigens is the most widely used phenotyping method. Recently, molecular typing (genotyping) has played a very important role in obtaining further discrimination when traditional typing (phenotyping) indicates close relationships between isolates, especially when the isolates are from a narrow geographical region or are obtained during a limited time. The correct interpretation of the molecular typing data is considered very important for providing epidemiological markers. These techniques are invaluable for epidemiological surveillance and monitoring of foodborne outbreaks, as they can be used to identify sources of contamination and determine the spread of the pathogen from food reservoirs to the food chain (Kumar et al., 2009). The Random Amplified Polymorphic DNA (RAPD) method, which employs 10 base pair random primers to locate random segments of genomic DNA to reveal polymorphisms, has been recommended as an alternative molecular fingerprinting tool. (Chansiripornchai et al., 2000; Khoodoo et al., 2002; Shabarinath et al., 2007).
The objectives of this study were to identify foodborne Salmonella spp. at the serotype level by both O and H antigen phenotyping, and to elucidate the utility of the RAPD method as a molecular genetic epidemiological tool. In addition, we aimed to establish baseline data on antibiotic-resistant Salmonella phenotypes in various types of retail foods obtained from free markets and supermarkets in Thailand.
Collection of retail food samples A total of 270 food samples were obtained comprising five categories of food: (1) raw meat samples (n = 51), which included 23 pork samples, 17 chicken meat samples and 11 beef samples; (2) leafy vegetable samples (n = 38) from independent farms and company farms that cultivate vegetables using organic and/or inorganic fertilizer; (3) fish or seafood (shrimp, shellfish and fish) samples (n = 37), which included sea and river fish; (4) fermented meat (n = 11), which included fermented ground pork with pork skins and fermented freshwater fish from lakes; and (5) tofu samples (n = 133), which included unpackaged tofu (n = 74) samples from free markets and packaged tofu (n = 59) samples from supermarkets. Tofu samples were purchased from twelve different free markets (3 in Bangkok, 4 in Pathum Thani, and 5 markets in Chiang Rai) and seven supermarkets (4 in Bangkok, 2 in Pathum Thani and 1 in Chiang Rai). All other food samples were purchased randomly from seven geographically different free markets and seven supermarkets in the Bangkok and Pathum Thani areas from September 2010 to February 2012. All samples were transported to a laboratory at Rajamangala University of Technology, Thanyaburi, within the shortest time possible in an insulated box with ice to maintain the temperature at 4 to 6°C. The pH values of the homogenized tofu samples in distilled water (1:2, wt/v) were measured using an electric pH meter (F-55 pH meter, Horiba, Kyoto, Japan).
Isolation Salmonella spp. were isolated as previously described (Ananchaipattana et al., 2012). A 25 g sample was homogenized with 225 mL of sterile Enterobacteriaceae Enrichment Mannitol (EEM) broth (Nissui Co., Ltd., Tokyo, Japan) and incubated at 35°C for 20 h. After incubation, 0.5 and 1.0 mL portions of the EEM-enriched sample were transferred to 10 mL of Rappaport-Vassiliadis (RV) broth and Hajna tetrathionate (TT) broth (Eiken, Tokyo, Japan), respectively. The RV and TT broth cultures were incubated at 42°C and 35°C, respectively, for 20 h. Following incubation, both cultures were streaked on Desoxycholate Hydrogen Sulfide Lactose (DHL) agar (Nissui), bismuth sulfide agar (Difco, New Hampshire, USA) and Mannitol Lysine Crystal violet Brilliant green (MLCB) agar (Nissui) an d incubated at 35°C for 20 h. Suspected colonies were picked and subjected to biochemical testing in Triple Sugar Iron (TSI) and Lysine Indole Motility (LIM) tubes and using the Salmonella LA “Seiken” latex-agglutination test (Denka-Seiken, Tokyo, Japan). Positive isolates were confirmed by API 20E (BioMerieux, Lyon, France).
Statistical analysis The differences in the number of positive samples among the groups of foods and between the two types of markets were evaluated with one-way analysis of variance (ANOVA) and Student's t-test, respectively. Differences were considered statistically significant at p-values less than 5%.
Identification of Salmonella spp. to the serovar level Salmonella serotypes were determined using Salmonella antiserum “Seiken” (Denka-Seiken, Tokyo, Japan). Each Salmonella isolate was grown overnight at 37°C on a nutrient agar plate (Nissui) and was then emulsified with 0.2 mL of 0.85% NaCl to prepare the somatic antigen suspension. Ten microliters of the suspension was locked on slide glass for 1 min with one drop of the respective somatic factor (O antiserum). Precipitation (a positive result) was detected and recorded. Flagella antigen detection was performed in Craigie tubes using brain-heart infusion broth (Nissui) with 0.3% agar. Cells were cultivated overnight in tryptic soy broth (Becton, Dickinson, Le Pont de Claix, France) with 0.6% yeast extract (Becton, Dickinson,). The flagella antigen suspension was prepared by adding 2 mL of 1% formalin in saline to 2 mL of an overnight culture in tryptic soy broth, then incubated at 37°C for 4 h prior to use. One drop of each flagellum factor (H antiserum) was placed in a glass tube, 0.2 mL of the suspension was added, and the mixture was incubated at 50°C for 1 h. This technique was applied to all Salmonella spp. in the study. Phase-2 flagella induction, if necessary, was performed according to the Kauffman-White serotyping scheme (Grimont and Weill, 2007).
DNA extraction Three of the 152 isolates of Salmonella spp. died during storage. Consequently, DNA was extracted from 149 isolated Salmonella spp. for RAPD fingerprinting. Single colonies of each of the 149 isolated Salmonella strains were transferred to 5 mL of brain-heart infusion broth and incubated at 37°C for 24 h. A 1 mL aliquot of the broth culture was centrifuged at 12,000 rpm for 5 min at 4°C, then the supernatant was discarded. The cells were suspended in 0.2 mL TE buffer, pH 8, mixed well on a vortex mixer, and boiled for 5 min at 95°C. The preparation was then centrifuged at 12,000 rpm for 5 min at 4°C. This process solubilized the DNA from the Salmonella cells into the supernatant, which was used for further studies. One microliter of the supernatant containing extracted DNA was used as the PCR template.
DNA analysis by RAPD-polymerase chain reaction (PCR) The reaction was conducted using six random primers, as show in Table 1. Fifty microliter PCR mixtures were made, each containing 25 µL of Premix Taq (Takara Bio, Shiga, Japan), 10 µM of the primer, and 20 ng of template DNA; the balance was sterile distilled water. Amplification was performed using the GeneAmp PCR 9700 system (Applied Biosystems, Foster City, CA, USA). The PCR conditions included an initial denaturation at 94°C for 5 min, followed by 45 cycles of denaturation at 94°C for 1 min, primer annealing at 35°C for 2 min, extension at 72°C for 2 min, and a final extension for 7 min at 72°C. The PCR products were kept at 4°C until further analysis. Ten microliters of PCR product were mixed with 2 µL of loading buffer (6X) and loaded on a 1.5% agarose gel, electrophoresed in TAE buffer, stained with ethidium bromide, then photographed and analyzed using a gel documentation system (ATTO, Tokyo, Japan). A score of ‘0’ for no visible band or ‘1’ for a visible band was awarded for the absence or presence, respectively, of a band for each profile obtained directly from the photographs of gels after electrophoresis and ethidium bromide straining. The scores for each primer were merged in a Microsoft Excel spreadsheet for further analysis. Genetic distance was calculated by simple matching and clustering by Ward's method (Batageli, 1988).
| Primer | Sequence (5'–3') | Reference |
|---|---|---|
| TS01 (PM5) | CGA CGC CCT G | Shabarinath et al., (2007) |
| TS02 (A) | AGC AGC GCC TCA | Miyata et al., (1995) |
| TS03 (OPI-06) | AAG GCG GCA G | Khoodoo et al., (2002) |
| TS04 (primer 1) | GGT GCG GGA A | Chansiripornchai et al., (2000) |
| TS05 (primer 2) | GTT TCG CTC C | Chansiripornchai et al., (2000) |
| TS06 (primer 3) | GTA GAC CCG T | Chansiripornchai et al., (2000) |
Antibiotic resistance assays All strains were tested for antibiotic resistance on Mueller-Hinton agar (Nissui) by the disk diffusion method (Bauer et al., 1966) using an SN-Disc (Nissui Pharmaceutical, Tokyo, Japan) according to the manufacturers' directions. The antimicrobial susceptibility of Salmonella isolates was judged by the diameter of the zone of inhibition from the antimicrobial disk. The sixteen antibiotics used in this experiment were ampicillin (ABPC) 10 µg, tazobactam/piperacillin (TAZ/PIPC) 10/100 µg, cefazolin (CEZ) 30 µg, cefuroxime sodium (CXM) 30 µg, cefoxitin (CFX) 30 µg, cefaclor (CCL) 30 µg, imipenem (IPM) 10 µg, azidothymidine (AZT) 30 µg, streptomycin (SM) 10 µg, kanamycin (KM) 30 µg, gentamicin (GM) 10 µg, tetracycline (TC) 30 µg, chloramphenicol (CP) 30 µg, polymyxin B (PL-B) 300 u, fosfomycin (FOM) 50 µg and nalidic acid (NA) 30 µg. The raw data are in the form of a zone size, which are interpreted based on the available CLSI (Clinical and Laboratory Standards Institute) data. The results for each Salmonella strain are reported as ‘susceptible’ or ‘resistant’.
The prevalence of Salmonella spp. in 152 food samples collected from two types of markets is shown in Table 2. Sixty-seven percent of meat samples tested were contaminated with Salmonella spp. In contrast, the percentage of vegetable (5%), fermented food (9%) or tofu (4%) samples contaminated with Salmonella spp. was comparatively low. Thirty-two serovars were identified among the 152 Salmonella spp. isolates. The top 10 serovars were S. Corvallis (n = 27), S. Rissen (n = 23), S. O4: i:- (n = 13), S. Kedougou (n = 10), S. Mbandaka (n = 8), S. Weltevreden (n = 7), S. Livingstone (n = 6), S. Agona (n = 5), S. Anatum (n = 5) and S. Hvittingfoss (n = 5). The two most commonly recovered serovars from chicken, pork and beef were S. Corvallis (52%: 17/33) and S. Kedougou (15%: 5/33), S. Rissen (14%: 7/52) and S. O4: i:- (14%: 7/52), and S. Rissen (27%: 6/22) and S. Livingstone (27%: 6/22), respectively. Only S. Hvittingfoss was identified from vegetable samples. The distribution of Salmonella spp. analyzed in this study is shown in Table 3.
| Categories of foods | Contaminated samples sold in | |||||
|---|---|---|---|---|---|---|
| free market | supermarket | total | ||||
| Number | (%) | Number | (%) | Number | (%) | |
| Meat* | 28/36 | 78 | 6/15 | 40 | 34/51 | 67 |
| Fish or seafood* | 7/17 | 41 | 1/20 | 5 | 9/37 | 24 |
| Vegetable | 1/18 | 6 | 1/20 | 5 | 2/38 | 5 |
| Fermented food | 1/4 | 25 | 0/7 | 0 | 1/11 | 9 |
| Tofu* | 5/74 | 7 | 0/59 | 0 | 5/133 | 4 |
| Total | 42/149 | 28 | 8/121 | 7 | 51/270 | 19 |
One hundred and forty-two (93%) Salmonella isolates were resistant to at least one antibiotic, 98 (64%) were resistant to three or more antibiotics, and 9 (6%) were resistant to 11 or more antibiotics. Salmonella isolates were most frequently observed to be resistant to streptomycin (78%), followed by tetracycline (59%) and ampicillin (51%). The serotype resistant to the largest number of antibiotics among the supermarket samples was S. Rissen (isolated from beef), which showed resistance to eleven of the sixteen antibiotics tested. For the free market samples, S. Corvallis (isolated from chicken) was resistant to thirteen antibiotics. The number and percentages of isolated-Salmonella serotypes showing antibiotic resistance are provided in Table 4.
The primers used for RAPD fingerprinting in this study are listed in Table 1. Genetic distance was calculated by simple matching and clustering by Ward's method (Batageli, 1988). Five groups of 149 isolated Salmonella strains were classified based on the results of RAPD fingerprinting (Fig. 1). Group 1 contains S. Corvallis; group 2 contains S. O4: i:- and S. Kedougou; group 3 contains S. Corvallis, S. Lexington, S. Give, S. Chester, S. Anatum, S. Worthington, S. Brunei, S. Mbandaka, S. Hvittingfoss, S. Agona, S. Weltevreden, and S. Hadar; group 4 contains S. O4: i:-, S. Typhimurium, S. Bardo, S. Stanley, S. ParatyphiB, S. Saintpaul, S. Ried, S. Orion, S. Yoruba, S. Krefeld, S. Amsterdam, S. Senftenberg, S. Kentucky, S. Virginia, S. Ohio, S. Muenchen, S. Glostrup and S. Livingstone; group 5 contains S. Rissen.
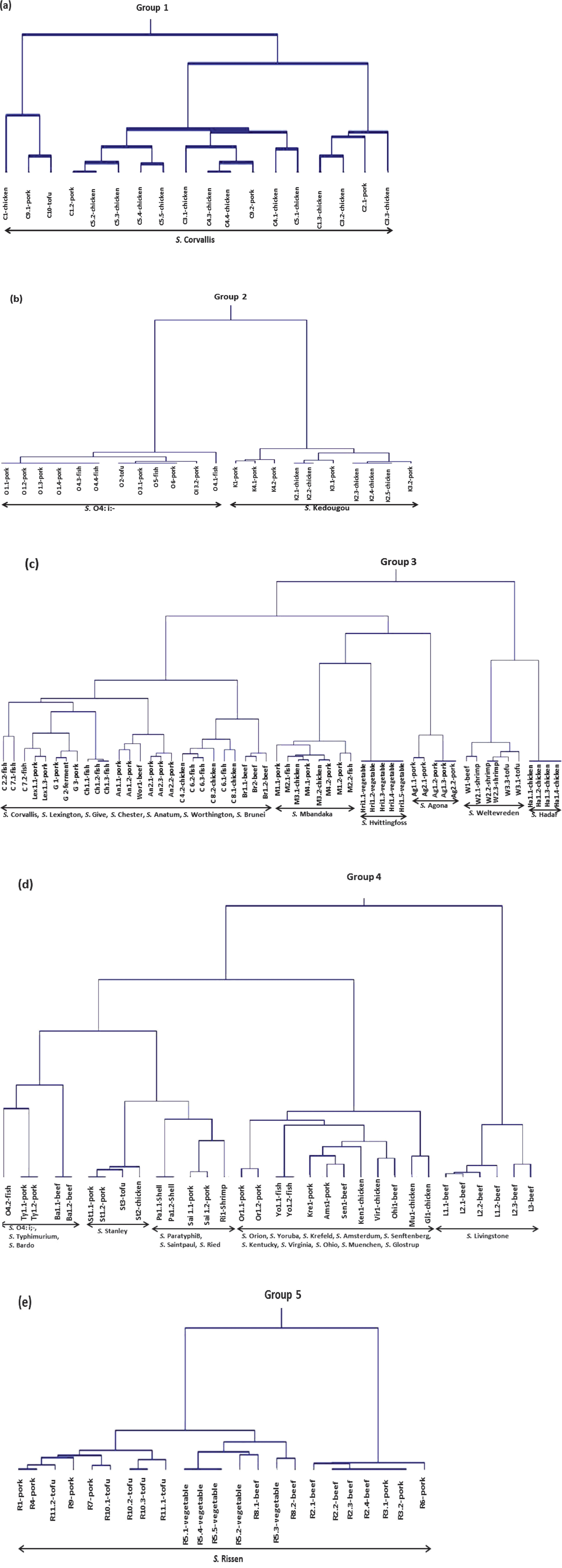
Dendrograms showing the five groups of the isolated Salmonella spp. (a) group 1, (b) group 2, (c) group 3, (d) group 4 and (e) group 5.
Salmonellosis remains a global challenge to public health. In Thailand, the incidence of foodborne disease has increased during the past ten years; however, the proportion of Salmonella infections has overall remained unchanged, despite some fluctuations (Ministry of Public Health, 2006). The results of this study show that the prevalence of Salmonella spp. in meat bought from free markets is higher than that in meat bought from supermarkets, which corroborates the findings of Vindigni et al. (2007). This difference in contamination level may reflect different risks of exposure to environmental contamination associated with different food distribution channels and food storage facilities. The majority of meat sold at free markets is processed in traditional slaughterhouses operated by meat vendors. The evisceration of chickens is usually done at the free market. There is no temperature control of the meat during processing and shipping to the market. In contrast, meats at supermarkets come from modern slaughterhouses operated by the supermarket company. In these modern facilities, the carcasses are hung for cutting and evisceration, then frozen before being cut into small er pieces and packaged for transport to domestic supermarkets (Padungtod et al., 2008).
The increasing antimicrobial resistance of Salmonella spp. has become a significant public health concern worldwide. Surveillance data demonstrated a noticeable increase in overall antimicrobial resistance among Salmonellae from 20% – 30% in the early 1990s to as high as 70% in some countries in the 2000s (Su et al., 2004). An increased level of resistance, especially to fluoroquinolones, has also been described in Thailand (Boonmar et al., 1998). Multidrug resistant Salmonella Schwarzengrund has spread from chickens to humans in Thailand and from imported Thai food products to humans in Denmark and the USA (Aarestrup et al., 2007). The present investigation indicated that 10 (7%) of the Salmonella isolates were sensitive to all of the antimicrobial agents tested, whereas nearly 93% of the isolates were resistant to at least one antibiotic and 64% were MDR, exhibiting resistance to three or more antibiotics. The antibiotics most commonly resisted by Salmonella were streptomycin, tetracycline, and ampicillin. These findings concur with previous reports that Salmonella isolates in retail food were highly resistant to tetracycline, streptomycin (Minami et al., 2010; Thong et al., 2011) and ampicillin (Piras et al., 2011).
Epidemiological investigations of Salmonella spp. provide important information about control measures that should be employed in the future. S. Enteritidis is one of the most common serovars that causes human salmonellosis in many countries (Omwandho and Kubota, 2010). Among 14,559 Salmonella isolates from chicken in Thailand, the most common isolate was S. Enteritidis (19.9%) (Bangtrakulnonth et al., 2004). Moreover, a ten-year study in Thailand indicated that S. Weltevreden was the most common serotype involved in human infections (Bangtrakulnonth et al., 2004; Padungtod and Kaneene, 2006). However, we isolated no S. Enteritidis from our retail food samples, and S. Weltevreden (3% of the Salmonella isolates) was a relatively uncommon serotype. The disparity between our findings and those reported previously might be due to differences in collecting time period, type of food sampled and number of samples. In this study, S. Rissen was a commonly found serotype, as previous studies have also found (Padungtod and Kaneene, 2006; Hendriksen et al., 2008). This Salmonella serovar is the most common serovar in Korea (Kim et al., 2011). The frequent occurrence of S. Rissen in water and food products indicates that a foodborne or waterborne reservoir is a possible source (Bangtrakulnonth et al., 2004). S. Corvallis has been reported to grow in chicken and pig meats (Archambault et al., 2006; Vindigni et al., 2007; Chuanchuen et al., 2008), which this study corroborates, as S. Corvallis was a commonly recovered serovar from chicken meat samples (52%). Historically, S. Anatum has been a major cause of salmonellosis in Thailand: it is found in a variety of animal sources (Bangtrakulnonth et al., 2004) and was the most common Salmonella serotype isolated from both beef and pork (Vindigni et al., 2007). In contrast, S. Anatum was not commonly recovered in the present study. S. Hvittingfoss was identified from only vegetable samples, suggesting that the occurrence of this serotype might be due to contamination from an environmental source.
The results of this and previous studies highlight the necessity to improve processing, distribution, storage conditions and temperature control to reduce the risk of widespread Salmonella spp. contamination of foods in both types of markets in Thailand. Furthermore, it is important to reduce the widespread use of antibiotics in animal feed in order to reduce the prevalence of antibiotic resistance in Salmonella spp.
RAPD analysis of 149 Salmonella isolates from retail foods from Thailand showed that they are genetically diverse. Six primers were selected for testing the samples by amplification. Most Salmonella serotypes were found in only one RAPD group (Fig. 1), with the exception of S. Corvallis and S. O4: i:-. S. Corvallis was found in group 1 and group 3. In group 1, S. Corvallis was isolated from chicken and pork, but the hosts of S. Corvallis in group 3 were mostly fish samples. S. O4: i:- was found in group 2 and group 4. In group 2, S. O4: i:- was isolated from pork, fish and tofu, but S. O4: i:- in group 4 was isolated from fish. The observed genetic diversity of S. Corvallis and S. O4: i:- may be due to the diversity of the host. The results of this study suggest that the RAPD method might be of value for screening and monitoring the epidemiology of Salmonella spp. Although the RAPD method shows poor repeatability (Mathis et al., 2011), the advantage of RAPD is that it can be used without previous knowledge of the nucleotide sequence of the target DNA; in addition, it is inexpensive and fast.
Acknowledgements This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (research project for ensuring food safety).