2017 Volume 23 Issue 5 Pages 679-688
2017 Volume 23 Issue 5 Pages 679-688
The prevalence of Staphylococcus aureus in dairy products in North Africa is high. Biopreservation promises to be one of the most sustainable strategies for reducing the risk of this pathogen. In this perspective, we screened local raw milk samples for LAB strains exhibiting anti-pathogenic activity against S. aureus and 11 other notorious food-borne pathogens. From the initial 120 LAB isolates, we selected the best-performing strains based on the breadth of their anti-pathogenic activity spectra. Lactobacillus paracasei subsp. paracasei KU517839 showed the best antibiosis against S. aureus with inhibition zones of (3.53 ± 0.05 cm) and the largest antibacterial activity spectrum as it inhibited all the other 11 pathogenic strains. Furthermore, a reduction of 5 log in S. aureus cell number was registered in fermented milk co-cultures and it significantly inhibited its growth in fresh cheese (8 log reduction; p < 0.001) even after 21 days of storage at 4°C.
Bio-preservation is a valuable and more sustainable strategy for preserving food quality and safety during handling and storage. Staphylococcus aureus is a leading cause of food-borne infections. Infection is induced by the ingestion of enterotoxins that are presecreted in food by virulent S. aureus strains. Once present in food, these enterotoxins A and D in particular are difficult to remove by conventional disinfection or sterilization techniques because they are heat stable and resistant to proteolytic enzymes (Forsythe, 2010). Biopreservation has the advantage of reducing the risk of their secretion in food in the first place. Tapping the potential of region-specific microflora for use as biopreservatives promises to discover more robust strains which are better adapted to local ecosystems. Furthermore, increasing consumer interest in healthy lifestyles, diets and concern about the possible adverse effects of chemically-preserved foods has compelled many food manufacturers to seek differentiation from competitors by offering more ‘natural’, ‘minimally-processed’ alternatives.
Algeria is the largest dairy consumer in the Maghreb (110 liters per capita per year for milk) and one of the main milk importers in the world (220 000 Metric Tons of milk powder in 2015) (USDA, 2015). Milk is considered as a staple food and the Algerian government has recently put in place various incentives in order to reduce imports, boost local production and encourage dairy processing units to use locally-produced milk rather than imported milk powder. However, many challenges face the national milk processors and one of them is the high variability of the hygienic quality of locally-produced milk. Available literature reports that milk from the Maghreb region has a microbial load which can be 100 fold more important than international standards, due to poor hygiene practices at farm level (Sraïri et al., 2013). Furthermore, it was reported that cow's milk produced by local farms is highly contaminated by Staphylococci (Mekademi et al., 2014; Hamiroune et al., 2014).
Milk pasteurization before human consumption or dairy products' manufacture is required in order to reduce the risk of foodborne infection. However, although pasteurization has decreased the initial microbial load of milk, it is not foolproof and does not yield a sterile product. Indeed, an initial population of 103 CFU/mL of S. aureus in milk may be sufficient for the production of enterotoxin A in cheese at detectable levels (García et al., 2007). Moreover, some traditional dairy fermented foods such as cheese and fermented milk are still prepared in Algerian households using raw milk. Biopreservation is a promising strategy in this regard, not only against S. aureus but also against other persistent food-borne pathogens which continue to pose a serious health threat worldwide. In this work, we screened lactic acid bacteria strains isolated from raw milk collected from locally-bred cows for their antibacterial activity against S. aureus as a first hurdle. The antibacterial activity spectra of the most performing strains were further characterized by challenging them with eleven (11) other bacterial food-borne pathogens. Following that, we validated the results by monitoring S. aureus cell growth in deliberately contaminated milk and fresh cheese inoculated with the best-performing strain.
Milk samples collection The collection of cow's milk was carried out between February and May 2013. Three samples of 250 mL each were collected from six farms (Kadir, Baaraouia, Meghezzi, Halmi, Oudar and Zadi) in the region of Constantine (Eastern Algeria), according to the standard AOAC (2002) method no 925.20. After washing the udder with bleach and wiping, the first streams were discarded and the following samples were collected and filtered before transporting them to the laboratory in a cooler at 4°C.
Isolation and phenotypic identification of lactic strains Isolation of LAB strains on various culture media was performed according to the methods described by FIL (1996). After incubation of the milk samples at 30°C to 45°C for 24 h, decimal dilutions of the milk samples in sterile peptone water (0.1% w/v; Sigma-Aldrich, USA) were performed. The LAB genera shown in Table 1 were isolated. The isolated bacteria were purified after several subcultures on the same agar medium. All strains were first subjected to macroscopic examination (color, size and shape of colonies), microscopic (morphology, cell regrouping mode and Gram stain), catalase test and the fermentation type. Gram-positive, catalase-negative and non-spore forming isolates were selected for physiological and biochemical identifications tests in accordance with other works (Bissonnette et al., 2000; González et al., 2007 and Azadnia et al., 2011). The profile of the fermentation of carbohydrates was determined using the API 50 CH galleries according to the manufacturer's instructions (bioMérieux, Marcy l'Etoile, France).
| Genera | Media (Sigma Aldrich, USA) | T (°C) | Duration (h) | Incubation |
|---|---|---|---|---|
| Lactococcus | Chalmers | 30 | 72 | Aerobiosis |
| Streptococcus | M17 | 45 | 72 | Aerobiosis |
| Leuconostoc | Elliker | 25 | 72–144 | Aerobiosis |
| Pediococcus | M17 | 30 | 72 | Aerobiosis |
| Mesophilic Lactobacillus | MRS | 30 | 24–36 | Anaerobiosis |
| Thermophilic Lactobacillus | MRS | 45 | 24–36 | Anaerobiosis |
Detection of antibacterial activity The 120 isolated strains were tested for their antagonism towards S. aureus ATCC 25923 by the agar spot test and the well diffusion assay as described by González et al. (2007). S. aureus ATCC 25923 was grown in trypticase soy broth (TSB; Sigma-Aldrich, USA) at 37°C for 24 h prior to experiments. For the agar spot test, 5 µL of overnight cultures of the isolated LAB strains were spotted onto the surface of MRS agar plates and incubated at 30 or 37°C for 24 h. A volume of 9 mL of semi-solid trypticase soy agar (TSA; Sigma-Aldrich, USA) containing 106 CFU/mL of S. aureus were poured over the plates of MRS agar containing the cultured LAB strains. The plates were incubated at 37°C for 24 h and monitored for the appearance of inhibition zones. The result was considered positive when the inhibition halo exceeds 0.2 cm in diameter.
Antibacterial activity was confirmed by a well diffusion assay. The test was performed using plates containing 106 CFU/mL S. aureus in TSA agar. Wells of 5 mm diameter were cut in and filled with 50 µL filtered supernatant fluid from MRS-cultured LAB strains. The plates were incubated at 37°C for 24 h. If antagonism was present, the possible inhibitory action of hydrogen peroxide was eliminated by adding a sterile solution of catalase (Sigma-Aldrich) at 1 mg/mL and leaving at 25°C for 30 min. If inhibition persisted, the presence of bacteriocins or bacteriocin-like compounds was considered likely. The proteinaceous nature of the active culture supernatant was evaluated using 4 proteolytic enzymes (Sigma-Aldrich, USA) separately at 1 mg/mL: Proteinase K (30 U/mg), α-Chymotripsin (40 U/mg), Trypsin (10 U/mg) and Protease from Streptomyces griseus (4 U/mg) (Bonade et al., 2001). The LAB which demonstrated the best activity anti S. aureus were selected to be tested against 11 pathogenic strains (Salmonella enterica ATCC 10708, Clostridium perfringens ATCC 13124, Yersinia enterocolitica ATCC 23715, Listeria monocytogenes ATCC 15313, Shigella sonnei ATCC 25931, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecium ATCC 19433, Citrobacter freundii ATCC 43864, Escherichia coli ATCC 25922, Aeromonas hydrophila ATCC 7966 and Vibrio parahaemolyticus ATCC 17802) by the agar spot test described above.
Genotypic identification of the antagonistic strains1) Extraction of genomic DNA
Total DNA of the antagonistic LAB strains was extracted from 5 mL pure cultures which were grown overnight in MRS broth at 30°C or 37°C by the method described by Leblond et al. (1996) with slight modifications. Bacterial cells were treated with: Tris-HCl (10 mM), EDTA (0.5 mM), lysozyme (50 mg/mL), proteinase K (20 mg/mL), SDS (10%) and potassium acetate (5 M) (Sigma-Aldrich, USA). DNA concentration was quantified by spectrophotometry using the nanodrop 2000c spectrophotometer (Thermo Scientific, USA).
2) PCR reaction protocols
PCR was carried out in a 50 µL mixture, containing 1 µL of bacterial genomic DNA, 23.5 µL of Emerald Amp GT PCR master mix kit TaKaRa RR310A (Takara, USA), 23.5 µL of sterile water, 1 µL (4 µM) of reverse primer rD1 (5′AAGGAGGTGATCCAGCC 3′) and 1 µL (4 µM) of forward primer fD1 (5′AGAGTTTGATCCTGGCTCAG 3′). Amplification of 16S rRNA gene was realized in a T100™ thermal cycler (Bio-Rad, USA), using the following amplification conditions: 35 cycles of 95°C for 1min, 55°C for 1min and 72°C for 7min. The first cycle was preceded by incubation for 5 min at 95°C. A volume of 5 µL of the PCR products and 8 µL of DNA-ladder were subjected to electrophoresis in a 1% agarose gel and were subsequently visualized by UV illumination after staining with SYBR Safe (Sigma-Aldrich, USA). Sequencing of the PCR products was performed by using the Sanger method (Eurofins Genomics, Germany). The results were analyzed by querying the database at the National Center for Biotechnology Information (NCBI). Data DNA sequences were deposited in GenBank with the following accession numbers: KU517835 (S29), KU517836 (S28), KU517837 (S27), KU517838 (S24), KU517839 (S10), KU517840 (S13), KU517841 (S9), KU517842 (S12), KU517843 (S11) and KU517844 (S35).
Verification of antagonism by co-culture in milk Overnight individual and co-cultures of the selected Lb. paracasei KU517839 and S. aureus ATCC 25923 were performed in TSB diluted with fresh sterile skimmed-milk and incubated at 37°C. The proportions used in the co-cultures were as follows: Lb. paracasei at 108 CFU/mL and S. aureus at 104 CFU/mL. The cell concentrations were monitored every three hours for 24 hours. Samples were removed and serially diluted into sterile saline solution. The dilutions were then plated on Chapman agar in triplicate, for S. aureus cells enumeration, and the average number of colonies obtained after 24 – 48 h incubation at 37°C was used to establish the growth and survival curves. The variation of pH was also monitored during the same incubation period and at the same intervals (Alomar et al., 2008).
Antagonism in fresh cheese1) Fresh cheese preparation
The flow diagram for preparation of the fresh cheese used in this study was identical to the process used by local Algerian cheese manufacturers. Briefly, 10 L of raw cow's milk were previously pasteurized at 72°C for 12 min, then cooled and maintained at a temperature of 30°C, enriched by 0.01 M of calcium chloride (Sigma-Aldrich, USA) in order to promote coagulation. The milk was subsequently divided into 2 batches of 5 L: the batch 1 was inoculated with Lb. paracasei KU517839 (108 CFU / mL) and S. aureus ATCC 25923 (104 CFU / mL), the batch 2 was inoculated with S. aureus ATCC 25923 (104 CFU / mL), used as control.
After milk reached a degree of acidification of 20 °Dornic, a quantity of rennet with a strength of 1/150000 (CHY-MAX®; Chr. Hansen, France) was added so that the coagulation time was brought to 15 min. Thereafter, manufacturing was conducted in the same manner for the two batches by cutting the curd followed by draining for 24 hours and then salting in brine (final concentration 15% w/v) for 15 min. The cheeses were then stored at 4°C until analysis. The experiment was repeated three times.
2) Sampling
Samples were taken during all phases of fresh cheese production. The first sample (1 mL of milk homogenized with 9 mL of a sterile tryptone-salt solution) was collected from each batch after inoculation with the lactic culture and S. aureus. After coagulation: 10 g of curd or cheese from each batch was homogenized with 90 mL of sterile tryptone-salt solution. After complete dissolution, decimal dilutions were made.
Growth of S. aureus (batches 1 and 2) and Lb. paracasei subsp. paracasei (batch 1) in the prepared cheeses was monitored via agar plate count on Chapman agar (Sigma-Aldrich, USA) and MRS respectively, every day during the first 5 days then on the 10th, 15th and 21st days of storage at 4°C for each cheese sample.
Statistical analysis All the experiments were performed in triplicate and results were expressed as mean ± standard deviation. Statistical analysis was performed using one-way analysis of variance (ANOVA) together with the Student's test using the software STATISTICA 10. Values of p < 0.05 were considered significant.
Isolation and identification of strains Out of a total of 265 isolated strains, 140 strains were Gram-positive, catalase negative and non-spore-forming. Twenty strains presumed enterococci were removed on the basis of their ability to grow at pH 9.6, 6.5% NaCl and at 45°C. The morphological, physiological and biochemical analyses of the remaining 120 strains revealed a diversity of LAB with 89 strains (74.2%) of them belonging to sphere-shaped LAB others 31 strains (25.8%) to the rod-shaped LAB which were classified on five presumptive genera (Table 2) : Lactobacillus (31 strains, 25.8%), Lactococcus (23 strains, 19.2%), Streptococcus (21 strains, 17.5%), Leuconostoc (30 strains, 25%) and Pediococcus (15 strains, 12.5%). Physiological and biochemical tests have allowed assignment of the strains to different species in the respective genera (Table 2). This diversity of genera was relative and dependent primarily on the nature of the material isolated and the different criteria used for each study, as reported by (Bissonnette et al., 2000; Badis et al., 2004; Guetouache et al., 2015a).
| Presumptive species | Growth at different temperatures (°C) | pNaCl (%)H | pH | Gas | Production of | Hydrolysis of | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 15 | 40 | 45 | 50 | RES | 2 | 3 | 4 | 6.5 | 4.2 | 4.5 | 4.8 | 6.5 | 7 | 8 | ADH | Acetoin | Dextran | Esculin | Citrate | ||
| Lactobacillus rhamnosus (7*) | + | + | − | + | + | + | + | − | − | + | ||||||||||||
| Lb. fermentum (6) | − | + | + | + | + | + | − | + | + | − | ||||||||||||
| Lb. plantarum (5) | + | − | − | + | + | + | + | − | − | + | ||||||||||||
| Lb. helveticus (5) | − | + | + | − | − | + | − | − | − | − | ||||||||||||
| Lb. brevis (4) | + | − | − | + | − | + | + | + | + | V | ||||||||||||
| Lb. casei subsp. casei (2) | + | V | − | + | + | + | + | − | − | + | ||||||||||||
| Lb. paracasei subsp. paracasei (2) | + | + | + | + | + | + | + | − | − | − | ||||||||||||
| Lactococcus lactis subsp. lactis (10) | + | + | − | V | + | + | − | − | + | − | + | − | + | − | ||||||||
| Lc. lactis subsp. cremoris (7) | + | − | − | − | + | − | − | − | + | − | − | + | V | − | ||||||||
| Lc. lactis biovar diacetylactis (5) | + | + | − | V | + | + | − | − | + | − | + | + | + | + | ||||||||
| Lc. raffinolactis (1) | + | − | − | − | + | − | − | − | − | − | V | + | + | V | ||||||||
| Streptococcus thermophiles (21) | − | + | + | + | V | − | − | − | + | − | − | − | − | |||||||||
| Leuconostoc mesenteroides subsp. mesenteroides (12) | + | − | − | − | − | − | + | + | − | + | − | − | ||||||||||
| Leu. pseudomesenteroides (8) | + | + | − | + | − | + | − | + | − | + | − | ± | ||||||||||
| Leu. lactis (6) | − | − | + | + | − | − | + | + | − | − | − | − | ||||||||||
| Leu. mesenteroides subsp. dextranicum (4) | + | + | − | ± | − | − | + | + | − | − | + | ± | ||||||||||
| Pediococcus acidilactici (11) | + | + | − | + | + | V | V | + | + | V | − | + | + | + | ||||||||
| Ped. damnosus (4) | − | − | − | − | − | − | + | − | − | − | − | − | V | − | ||||||||
(−) Less than 10% of reaction positive, (+) more than 90% of positive reactions, (V) more than 10% and less than 90% of positive reactions, GAS—production of gas from glucose, ADH—arginine dihydrolase, RES—heat resistance at 63.5°C during 30 min.
Antibacterial activity Among the 120 strains studied, only 35 strains have shown a significant antibiosis against S. aureus ATCC 25923 using the agar spots test. The 35 strains were further tested by the agar well diffusion assay. Except for 4 strains, culture supernatants of all other 31 strains exerted an anti-S. aureus activity. After exclusion of the inhibition due to organic acids and hydrogen peroxide, only 10 strains have shown antagonism towards S. aureus (Table 3).
| Average diameter of inhibition zone±S.D (cm) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Agar spot tests | Agar well diffusion assays | |||||||
| Supernatant without any treatment | Supernatant with pH neutralized to 6.5 | Supernatant with pH neutralized to 6.5 and H2O2 eliminated | Relative activity of the supernatants after treatment with enzymes | |||||
| Proteinase K | α-Chymotripsin | Trypsin | Protease from Streptomyces griseus | |||||
| Strains | Lactobacillus | |||||||
| S9 (KU517841) | 3.10±0.10b | 1.08±0.01d | 0.84±0.01c | 0.81±0.01b,c | − | − | − | − |
| S10 (KU517839) | 3.53±0.05a | 1.68±0.02a | 1.21±0.02a | 1.10±0.05a | − | − | − | − |
| S11 (KU517843) | 3.18±0.07b | 1.43±0.05b | 0.85±0.05c | 0.81±0.05b,c | − | − | − | − |
| S12 (KU517842) | 2.83±0.15c,d | 1.25±0.05c | 0.63±0.07d,e | 0.61±0.05d | − | − | − | − |
| S13 (KU517840) | 2.66±0.15e,f | 1.10±0.10d | 0.60±0.08e | 0.58±0.10d | − | − | − | − |
| Leuconostoc | ||||||||
| S24 (KU517838) | 2.86±0.05c | 1.33±0.05b,c | 1.07±0.02b | 1.07±0.02a | − | − | − | − |
| S27 (KU517837) | 2.70±0.10d,e | 1.03±0.02d | 1.02±0.02b | 1.01±0.01a | − | − | − | − |
| S28 (KU517836) | 2.76±0.05c,d,e | 1.23±0.05c | 0.90±0.10c | 0.90±0.10b | − | − | − | − |
| S29 (KU517835) | 2.53±0.05f | 1.03±0.15d | 0.65±0.05d,e | 0.65±0.05d | − | − | − | − |
| Pediococcus | ||||||||
| S35 (KU517844) | 2.76±0.05c,d,e | 1.33±0.05b,c | 0.75±0.05d | 0.75±0.05c | − | − | − | − |
Values are means ± standard deviations of triplicates.
(−) Represents no inhibition zone.
Lactobacillus (S10) showed the strongest antibiosis against the indicator strain with the largest zone of inhibition (3.53 ± 0.05 cm) (p < 0.05). The antibacterial activity of these 10 strains was eliminated after treatment of the active supernatants with various proteolytic enzymes (Proteinase K, α-Chymotripsin, Trypsin and Protease from Streptomyces griseus) which indicates implication of peptides or proteins in the activity.
The results of the antagonism test of the selected 10 LAB strains against 11 pathogenic bacteria are shown in Table 4. The inhibitory activity against Clostridium perfringens ATCC 13124 was the lowest in all ten LAB strains tested. Some strains did not show any significant inhibition activity against the indicator pathogenic strains: Clostridium perfringens ATCC 13124 (Lactobacillus S13), (Leuconostoc S24 and S27), Pseudomonas aeruginosa ATCC 27853 (Leuconostoc S27) and Enterococcus faecium ATCC 19433 (Leuconostoc S24).
| Pathogens | Strains of lactic acid bacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S9 (KU517841) | S10 (KU517839) | S11 (KU517843) | S12 (KU517842) | S13 (KU517840) | S24 (KU517838) | S27 (KU517837) | S28 (KU517836) | S29 (KU517835) | S35 (KU517844) | |
| S. enterica ATCC 10708 | 1.07±0.025d | 1.79±0.03a | 1.16±0.11c | 1.02±0.02d | 1.01±0.01d | 1.44±0.05b | 0.57±0.02f | 0.92±0.03e | 1.42±0.03b | 1.05±0.02d |
| C. perfringens ATCC 13124 | 0.75±0.02b | 0.93±0.03a | 0.90±0.03a | 0.92±0,02a | 0±0d | 0±0d | 0±0d | 0.6±0.01c | 0.8±0.01b | 0.72±0.02b |
| Y. enterocolitic ATCC 23715 | 1±0.01d,e | 2.33±0.03a | 0.91±0.01e,f | 1.06±0.02d | 1.02±0.01d,e | 2.16±0.15b | 0.78±0.01f | 1.5±0.02c | 1.44±0.03c | 1.05±0.03d,e |
| L. monocytogenes ATCC 15313 | 1.01±0.01d | 1.92±0.02a | 1.35±0.05b | 1.23±0.04b,c | 1.04±0.03d | 1.06±0.05d | 0.61±0.01e | 1.04±0.01d | 1.34±0.04b | 1.1±0.01c,d |
| S. sonnei ATCC 25931 | 1.29±0.01d,e | 2.75±0.02a | 1.16±0.05g | 1.25±0.01e,f | 1.25±0.01f | 1.51±0.01c | 0.7±0.01h | 1.16±0.01g | 1.91±0.01b | 1.31±0.01d |
| P. aeruginosa ATCC 27853 | 1.19±0.01f | 1.61±0.01a | 1.41±0.01b | 1.17±0.01g | 1.12±0.02h | 1.31±0.01c | 0±0j | 1.22±0.01e | 1.26±0.01d | 0.87±0.01i |
| E. faecium ATCC 19433 | 1±0.00f | 1.45±0.02b | 1.13±0.01e | 1.28±0.01c | 0.88±0.01g | 0±0j | 0.6±0.01i | 1.51±0.01a | 1.19±0.00d | 0.69±0.01h |
| C. freundii ATCC 43864 | 1.16±0.01f | 1.42±0.01d | 1.55±0.02b | 1.15±0.01f | 1.05±0.01g | 1.49±0.01c | 0.69±0.01i | 1.77±0.01a | 1.23±0.01e | 0.84±0.01h |
| E. coli ATCC 25922 | 1.04±0.01g | 2.52±0.02a | 1.3±0.02e | 1.16±0.01f | 1.09±0.00f,g | 2.3±0.1b | 0.62±0.01h | 1.24±0.02e | 1.71±0.01d | 1.78±0.01c |
| A. hydrophila ATCC 7966 | 0.8±0.01g | 1.6±0.02a | 1.52±0.02b | 1.29±0.01d | 1.35±0.01c | 1.28±0.01d,e | 0.82±0.03g | 1.25±0.01e | 0.72±0.4h | 0.93±0.01f |
| V. parahaemolyticus ATCC 17802 | 1.01±0.00e | 1.8±0.01a | 1.02±0.01d,e | 1.03±0.01d | 1.01±0.00e | 1.6±0.01b | 0.62±0.01g | 1.6±0.01b | 1.17±0.02c | 0.91±0.01f |
Values are means ± standard deviations of triplicates.
Lactobacillus S10 had the broadest spectrum of inhibition against all 11 indicator bacteria with the highest inhibition zone diameters (p < 0.05). This strain was therefore selected for further tests in skimmed milk and fresh cheese co-cultures.
Identification of the antagonistic LAB strains using 16S rRNA gene sequencing The results of sequencing of the 16S rRNA gene sequences for the 10 LAB strains identified the isolates as follows: Leuconostoc mesenteroides subsp. mesenteroides KU517838 (S24), Leuconostoc mesenteroides subsp. mesenteroides KU517836 (S28), Leuconostoc mesenteroides subsp. mesenteroides KU517835 (S29), Lactobacillus fermentum KU517841 (S9), Lactobacillus fermentum KU517842 (S12), Lactobacillus fermentum KU517840 (S13), Leuconostoc pseudomesenteroides KU517837 (S27), Lactobacillus rhamnosus KU517843 (S11), Lactobacillus paracasei subsp. paracasei KU517839 (S10) and Pediococcus acidilactici KU517844 (S35).
Co-cultures in skimmed milk The results reported in Fig. 1 show that S. aureus ATCC 25923 in pure culture presented a normal growth curve, with an exponential growth phase from the beginning of the culture. In the case of pure cultures, we registered an increase in the concentration of cells to 7.7 × 108 CFU/mL and the stabilization of the pH at 6.6 (Fig. 2) after an incubation period of 24 h. As for co-cultures, the count of S. aureus decreased significantly (p < 0.05) between 9 h and 24 h to attain 103 CFU/mL, compared to the pure culture (5 log lesser). Furthermore, the pH decreased to 3.7 after 24 h incubation. The decrease of S. aureus count could be explained by the antagonistic effect of the Lb. paracasei subsp. paracasei strain towards S. aureus; this could be the result of the synthesis of inhibitory metabolites as demonstrated in the well diffusion assay (Alomar et al., 2008; Scatassa et al., 2015).
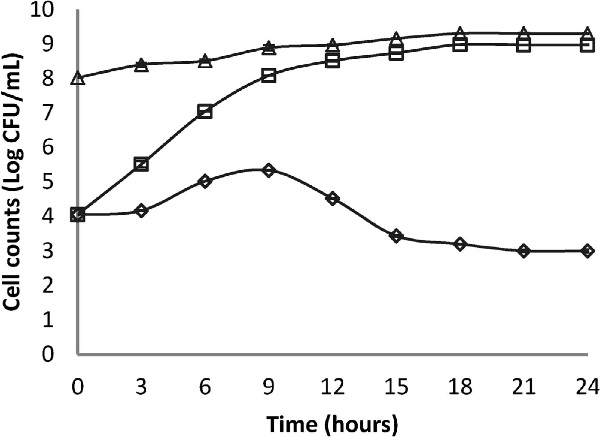
Changes in counts of S. aureus (⋄), Lb. paracasei (▵) in skimmed milk in co-cultures (104 CFU/mL of S. aureus + 108 CFU/mL of Lb. paracasei) and of S. aureus in pure cultures (S. aureus at 104 CFU/mL) (□).

Changes in pH values of sterile skimmed milk in pure cultures (S. aureus at 104 CFU/mL) (□) and in co-cultures (104 CFU/mL of S. aureus + 108 CFU/mL of Lb. paracasei) (⋄).
Our results are in agreement with several similar works which identified endemic strains from Lactobacillus species as either the most active or having the widest antagonism spectrum compared to other LAB genera. For instance, Guetouache et al. (2015b) performed a screening of antibacterial activity against S. aureus ATCC 6538 of 29 LAB strains isolated from a traditional Algerian cheese, by the agar well diffusion assay and identified 6 bacteriocin-like producing strains. Bendali et al. (2011) studied the antagonism of Lactobacillus paracasei subsp. paracasei against S. aureus in fermented milk and registered a reduction of 2 log of S. aureus in co-cultures after 24 h incubation. Several studies on the use of Lactobacillus strains for the production of fermented foods including dairy products (fermented milks and cheeses) noted the important role that these bacteria have in the biopreservation of these products against alteration and pathogenic bacteria like; S. aureus, S. typhi, L. monocytogenes, B. cereus, E. coli and S. flexneri (Léonard et al., 2015; Júnior et al., 2015; Seyed and Bagher, 2016).
Scientific literature abounds with reports about the biopreservation properties of LAB. These properties are due to the synthesis of a wide range of products from low molecular mass compounds, such as hydrogen peroxide, carbon dioxide and diacetyl, to high molecular mass compounds, such as bacteriocins. Organic acids produced by LAB lead to a reduction in pH levels. These products exhibit antibacterial activity against various pathogenic microorganisms, including Gram-positive and Gram-negative bacteria (Favaroa et al., 2015; Ganzorig et al., 2016).
Antagonism in fresh cheese The results reported in Fig. 3 show that the number of S. aureus in the control cheese (batch 2) increased steadily from the 1st day of storage, with a population of 8.9 log and continued to rise until the 4th day when it reached a maximum population of 9.4 log. From the 10th day, a plateau was reached with a bacterial load of 8 log which was maintained until the 21st day of storage.
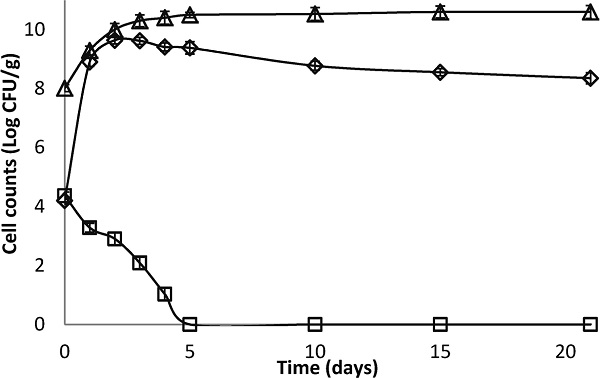
Changes in counts of S. aureus in freshly made cheese in the presence and in the absence of Lb. paracasei. (□): batch 1 (4 log CFU/mL of S. aureus + 8 log CFU/mL of Lb. paracasei), (⋄): batch 2 (S. aureus at 4 log CFU/mL). (▵): changes in counts of Lb. paracasei in freshly made cheese in the batch 1.
In batch 1, a markedly significant difference (p < 0.05) in the number of S. aureus was observed from day 1, compared to the control batch. The S. aureus count continued to decline until it reached the lethal phase on the 5th day where it stabilized until the 21st day of storage.
With regards to the growth of Lb. paracasei subsp. paracasei, it reached 10.5 log on the 5th day of storage and stabilized thereafter at 10.6 log from the 15th till the 21st day of storage. The significant inhibition (p < 0.05) of S. aureus in cheese (batch 1) compared to the control may be due to the synthesis of anti-staphylococcal substances in the cheese matrix by Lb. paracasei subsp. paracasei (Georgievaa et al., 2015; Castillo et al., 2015). Such substances are well referenced in the scientific literature. For instance, Pinto et al. (2011) studied the inhibition of S. aureus by nisin in a traditional cheese (Minas Serro) and reported a 2 log reduction in S. aureus count after 7 days maturing and a 3 log reduction after 45 days of ripening.
Based on our results, we can reasonably conclude that Lb. paracasei subsp. paracasei KU517839 is a promising strain for potential use as a biopreservative agent against S. aureus in cheese, particularly cheese made from pasteurized milk (Heredia-Castro et al., 2015; Yoon et al., 2016). Several other independent research studies have shown that the species Lb. paracasei could be used as a probiotic and as biopreservative agent in food (Jeon et al., 2016; Mahmoudia et al., 2016).
In this work, we report the screening and identification of 10 bacteriocin-like producing LAB strains from 120 strains isolated from Algerian cow's milk. The isolates were selected based on their best and widest antagonistic activity against a panel of 12 food-borne pathogens. The selected strains were identified as 3 strains of Leuconostoc mesenteroides subsp. mesenteroides, 3 strains of Lactobacillus fermentum, 1 strain of Leuconostoc pseudomesenteroides, 1 strain of Lactobacillus rhamnosus, 1 strain of Lactobacillus paracasei subsp. paracasei and 1 strain of Pediococcus acidilactici. Lactobacillus paracasei subsp. paracasei showed the best performance against all 12 indicator pathogenic strains tested. It resulted in a reduction of 5 log in S. aureus ATCC 25923 cell numbers in fermented milk co-cultures, it also resulted in a 100% inhibition of S. aureus in freshly made cheese after 5 days of storage in comparison with respect to the control batch. Our results showed the potential of autochtonous LAB strains for use as biocontrol agents in locally manufactured dairy foods. Biopreservation is a more sustainable strategy for combatting food-borne infections, especially in rural settings or in manufacturing facilities and supply-chains which lack appropriate quality assurance technologies.
Aknowledgements This study was conducted with the financial support of the Direction Générale de la Recherche Scientifique et du Développement (DGRSDT), Ministry of High Education and Scientific Research, Algeria. Our grateful thanks to Abdellah Zikiou, Bachir Benkaddour, Asma Meghezzi, Abderrahmane Selmania and Assia Ikhlef from the Biotechnology Research Center (C.R.Bt) for skilful technical assistance. Our grateful thanks to Frédéric Borges, Anne-Marie Revol-Junelles and Stéphane Désobry from the Laboratoire d'Ingénierie des Biomolécules (LIBio), E.N.S.A.I.A., Université de Lorraine, France, for hosting us in their lab and facilitating the completion of the molecular identification of the strains.