2019 Volume 25 Issue 1 Pages 19-27
2019 Volume 25 Issue 1 Pages 19-27
To develop a method for the identification of shiikuwasha (Citrus depressa Hayata) and calamondin (Citrus madurensis Lour.), trnL-trnF and trnT-trnL intergenic spacer regions of their chloroplast DNA were amplified using PCR and the nucleotide sequences were determined. In each region, a single nucleotide polymorphism (SNP) site specific to the respective citrus species (shiikuwasha and calamondin) was found. For species discrimination using PCR, two forward primers containing the allele-specific SNP site at the 3′-end and a mismatched nucleotide at the 3rd base from the 3′-end were designed. The allele-specific forward primers specific to shiikuwasha and calamondin were respectively designated CiDeLF-F and CiMaTL-F. To confirm the specificity of the designed primers, PCR was carried out with DNA prepared from citrus peel or hand-squeezed juice as the template. Results showed that shiikuwasha and calamondin fruits and juices were identifiable by PCR using the allele-specific primers. Furthermore, this allele-specific PCR method can be applied to industrially processed and concentrated juice by amplifying DNA in advance.
Shiikuwasha (Citrus depressa Hayata) is one of the citrus species and is a very popular fruit in the northern areas of Okinawa main island, Japan. Several cultivars are categorized as shiikuwasha; of these, ‘Ogimi kugani’, ‘Katsuyama kugani’, ‘Izumi kugani’ and ‘Kaachi’ are the most commonly cultivated. Shiikuwasha is characterized by a sour taste and distinctive flavor. Most of the harvested fruit is processed into beverages and consumed as juice. The fruit contains high levels of polymethoxylated flavones (Nogata et al., 2006). Of these flavones, nobiletin is reported to show antitumor effects against gastric cancer, antiobesity activities, hepatoprotective effects against D-galactosamine-induced liver injury and improvement of memory impairment (Minagawa et al., 2001; Lee et al., 2011; Akachi et al., 2010; Onozuka et al., 2008). Therefore, shiikuwasha has been recognized as a health-promoting food and demand for the fruit has risen, leading to increases in the market price of shiikuwasha fruits and a shortage of raw materials. As a result, calamondin (Citrus madurensis Lour.) juice, an inexpensive ingredient, has been used to deliberately adulterate processed shiikuwasha products (juice) by manufacturers, leading to the circulation of fake products. Calamondin is widely distributed in China, the Philippines, the United States, and Japan (Moshonas et al., 1996). According to Tanaka's classification (Tanaka, 1969), shiikuwasha and calamondin belong to Citrus-Metacitrus-Acrumen and Citrus-Metacitrus-Pseudofortunella, respectively; thus, these citruses are considered to be taxonomically different species. The fruits can be distinguished visually. However, if they are once juiced and mixed, it is difficult to identify the mixing of calamondin juice to shiikuwasha juice without scientific analysis.
As examples of fruit juice adulteration, the addition of water to juice, excessive dilution of concentrates, additions of sugar, pulp wash (re-eluate from the addition of water to residual pulp), or substitution of cheap ingredients have been reported (Le Gall et al., 2001; Widmer et al., 1992). Thus, analytical techniques are required to discriminate processed products such as juice. Of the discrimination methods, DNA-based polymerase chain reaction (PCR) methods are currently used for authentication of various foods such as rice, olive oil, fruits, wine and orange juice (Otsubo et al., 2005; Breton et al., 2004; Siret et al., 2000; Aldeguer et al., 2014).
In particular, single nucleotide polymorphisms (SNPs), which appear at a rate of one per 100 – 1,000 base pairs (bp) in genomic DNA (Ching et al., 2002), have been utilized as genetic markers (Aldeguer et al., 2014; Kitaoka et al., 2010; Pardo, 2015). SNP analysis can reliably distinguish specific bases and be applied to identify samples with advanced decomposition such as juice or those containing closely related varieties. Allele-specific PCR methods including SNPs have been developed (Newton et al., 1989; Okayama et al., 1989) and utilized for discrimination of alleles (Kwok et al., 1990; Drenkard et al., 2000).
In order to develop and apply allele-specific PCR techniques to discriminate between shiikuwasha and calamondin, data of regions including SNPs specific to the species are vital. For this purpose, non-coding sequences of two intergenic spacer regions, between the trnL (tRNA-Leu) and trnF (tRNA-Phe) genes and the trnT (tRNA-Thr) and trnF genes, were selected as target regions. These regions are characterized by relatively high rates of nucleotide substitution and have demonstrated utility in phylogenetic relationship studies (Taberlet et al., 1991). Polymorphism in the trnL-trnF intergenic spacer region was used for detection of pine hybrids (Chen et al., 2002) and identification of a type of cactus (Aragane et al., 2011).
In the present paper, we determined the nucleotide sequences of the trnL-trnF and trnT-trnF intergenic spacer regions of chloroplast DNA to develop newly designed PCR primers including the corresponding SNP sites. Then, application of the designed primer sets for identification of shiikuwasha and calamondin was investigated.
Materials Fruits of shiikuwasha (C. depressa Hayata) cultivars, ‘Izumi kugani’, ‘Ogimi kugani’, ‘Katsuyama kugani’, ‘Tozanwase’, ‘Kaachi’, ‘B-41’, ‘B-41-H’, and of calamondin (C. madurensis Lour.) were obtained from the Okinawa Prefectural Agricultural Research Center (Nago, Okinawa, Japan) and Taiwan Agricultural Research Institute, Council of Agriculture, Executive Yuan (Taichung, Taiwan), during the 2008 harvest season. Whole fruits of shiikuwasha and calamondin were respectively squeezed into juice using a 260 mm long hand-squeezed juicer. The obtained juice was designated as hand-squeezed juice. Concentrated shiikuwasha and calamondin juices, which had been industrially prepared, were respectively obtained from companies in Japan and Taiwan (Table 1). Fruit samples were peeled, and then the peels and pulps were stored separately in the freezer at −80 °C. Hand-squeezed and concentrated juices were also stored at −80 °C freezer until use.
| Sample name, type | Production areas | Scientific name | Common name | DDBJ accession number | |
|---|---|---|---|---|---|
| trnL-trnF | trnT-trnL | ||||
| Izumi kugani, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377852 | LC377861 |
| Ogimi kugani, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377853 | LC377862 |
| Katsuyama kugani, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377854 | LC377863 |
| Tozanwase, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377855 | LC377864 |
| Kaachi, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377856 | LC377865 |
| B-41, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377857 | LC377866 |
| B-41-H, peel | Okinawa | C. depressa Hayata | Shiikuwasha | LC377858 | LC377867 |
| Calamondin, peel | Okinawa | C. madurensis Lour. | Calamondin | LC377859 | LC377868 |
| Calamondin, peel | Taiwan | C. madurensis Lour. | Calamondin | LC377860 | LC377869 |
| Shiikuwasha, peel | Taiwan | C. depressa Hayata | Shiikuwasha | – | – |
| Hand-squeezed shiikuwasha juice | Okinawa | C. depressa Hayata | Shiikuwasha | – | – |
| Hand-squeezed calamondin juice | Okinawa | C. madurensis Lour. | Calamondin | – | – |
| Concentrated shiikuwasha juice | Okinawa | C. depressa Hayata | Shiikuwasha | – | – |
| Concentrated calamondin juice | Taiwan | C. madurensis Lour. | Calamondin | – | – |
DNA preparation from citrus peel The collected citrus peels were freeze-dried, and then crushed and ground using a homogenization pestle. Total DNA was prepared using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's protocol.
DNA preparation from juice and amplification of purified DNA DNA was purified from hand-squeezed juices and concentrated juices using a DNeasy Plant Mini Kit (Qiagen). The purified DNA was then amplified using an Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare UK Ltd., Little Chalfont, Buckinghamshire, UK).
Measurement of DNA concentration Template DNA was prepared from total DNA of citrus peel. The concentration and purity of template DNA were determined by UV absorbance (NanoDrop ND-1000; NanoDrop Technologies Co., Ltd., Wilmington, DE, USA).
Amplification of trnL-trnF and trnT-trnL regions The trnL-trnF and trnT-trnL intergenic spacer regions were subjected to PCR amplification according to the methods of Taberlet et al. (1991). For amplification, the primers used were as follows: trnL-trnF forward, 5′-GGTTCAAGTCCCTCTATCCC-3′; trnL-trnF reverse, 5′-ATTTGAACTGGTGACACGAG-3′; trnT-trnL forward 5′-TCTACCGATTTCGCCATATC-3′; trnT-trnL reverse, 5′-CATTACAAATGCGATGCTCT-3′. The PCR mixture contained 5 ng of template DNA, 14.5 µL of sterile distilled water, 2.5 µL of 10× Ex Taq buffer, 2 µL of 25 mM MgCl2, 2 µL of 2.5 mM dNTP mixture, 0.5 µL of each primer (10 pmol/µL), and 5 units of Ex Taq polymerase (Takara Bio Inc., Shiga, Japan). The amplification conditions were as follows: 1 cycle of 5 min at 95 °C; 35 cycles of 1 min at 92 °C, 1 min at 55 °C, 1 min at 72 °C; and a final cycle of 5 min at 72 °C. Amplification was performed in a programmable temperature control system (Astec Co., Ltd., Fukuoka, Japan).
PCR products were loaded onto a 2 % agarose gel in 1×TAE buffer solution, stained with ethidium bromide and photographed under UV light.
DNA sequencing and alignment of sequences PCR products were purified using a QIAquick PCR purification kit, according to the manufacturer's protocol (Qiagen). Eighteen purified PCR products were directly sequenced using an ABI Prism Dye Terminator Cycle Sequencing Ready Kit with AmpliTaq DNA polymerase (Thermo Fisher Scientific Inc., Kanagawa, Japan). For DNA sequencing of the trnL-trnF intergenic spacer region, the same primer pairs for PCR amplification were used. On the other hand, for the trnT-trnL intergenic spacer region, new primers were designed based on the nucleotide sequence of Citrus sinensis (DDBJ Accession No.DQ864733). The primers used were trnT-trnL forward for sequencing, 5′-GTCTAAATC GAATATTATATTCGATTC-3′; trnT-trnL reverse for sequencing, 5′-GAATACTTCGAACGGTCGATT-3′. The determined nucleotide sequences of the trnL-trnF and trnT-trnL regions from individual citrus cultivars were respectively aligned using the CLUSTALW program (http://www.genome.jp/tools-bin/clustalw) and compared for identification of SNP sites.
Design of allele-specific primers In order to detect species-specific SNPs in the trnL-trnF and trnT-trnL intergenic spacer regions, allele-specific PCR was utilized as described by Okayama et al. (1989). For this purpose, a forward primer with a species-specific base at the 3′-end corresponding to the SNP site was designed. In addition, the third base from the 3′-end was substituted with a mismatch base in order to reduce nonspecific amplification (Hayashi et al., 2004). Each primer is shown in Table 2.
| Location | Expected size | Name | Sequence |
|---|---|---|---|
| Shiikuwasha | 76 bp | CiDeLF-F | 5′-CTCCTACCTTCTCCTTTTaTT-3′ |
| trnL-trnF | CiDeLF-R | 5′-GGAAATGGAAAAGAGTAGGATAG-3′ | |
| Calamondin | 105 bp | CiMaTL-F | 5′-GCTATTATATTCTAAATGATTAATAAAtAC-3′ |
| trnT-trnL | CiMaTL-R | 5′-CCTTAAGGAAGAACCTATAT-3′ |
The underlined nucleotide indicates SNP site specific to each citrus. The nucleotide in small capital indicates substituted mismatch base.
Allele-specific PCR The PCR mixture contained 5 ng/µL of template DNA, 1.4 µL of sterile distilled water, 5 µL of 2×PCR buffer, 2 µL of 2 mM dNTP mixture (Toyobo), 0.2 µL of each primer (10 pmol/µL) and 1 unit of Ex Taq polymerase (Toyobo) and was placed on ice. The amplification parameters were as follows: 1 cycle of 2 min at 94 °C; 35 cycles of 10 sec at 98 °C, 30 s at between 54.9 °C and 65 °C, 30 s at 68 °C; and a final cycle of 10 min at 68 °C.
PCR amplification and determination of nucleotide sequences of trnL-trnF and trnT-trnL intergenic spacer regions DNAs prepared from seven shiikuwasha and two calamondin peels, respectively, were used for amplification of the trnL-trnF and trnT-trnL intergenic spacer regions. As shown in Fig. 1A, the size of PCR products amplified with the trnL-trnF primers was approximately 450 bp, which was almost the same as the expected size of the trnL-trnF region (437 bp). In Fig. 1B, the size of amplified PCR products with the trnT-trnL primers was about 1,100 bp; as the expected size was 1,121 bp, successful amplification of the trnT-trnL region was indicated. Then, we determined the nucleotide sequences of the PCR products of the trnL-trnF and trnT-trnL regions. The nucleotide sequences of the regions were deposited at DDBJ under accession numbers from LC377852 to LC377869 (Table 1).
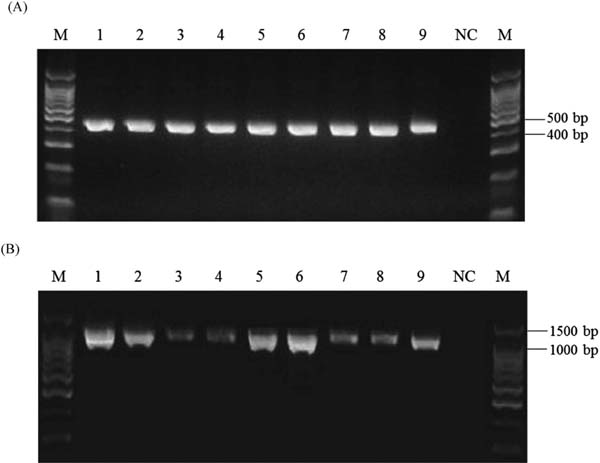
Amplification of trnL-trnF (A) and trnT-trnL (B) intergenic regions of shiikuwasha and calamondin. Lanes: M, 100 bp DNA ladder; 1, Izumi kugani; 2, Ogimi kugani; 3, Katsuyama kugani; 4, Tozanwase; 5, Kaachi; 6, B-41; 7, B-41-H; 8, calamondin from Okinawa; 9, calamondin from Taiwan; NC, negative control (without DNA template).
The determined nucleotide sequences were compared to the registered whole genome of orange (C. sinensis) (DQ864733) using DDBJ. As shown in Fig. 2, the determined nucleotide sequences were highly similar to those of the target (trnL-trnF and trnT-trnL) regions and the nucleotide sequences were highly conserved between shiikuwasha and calamondin. Although there were few differences in the nucleotide sequences of both regions between shiikuwasha and orange, calamondin had two specific SNPs in the trnT-trnL and the trnL-trnF regions corresponding to the 50268th position and the 51403th position of reference sequence (DQ864733.1) of orange, respectively.
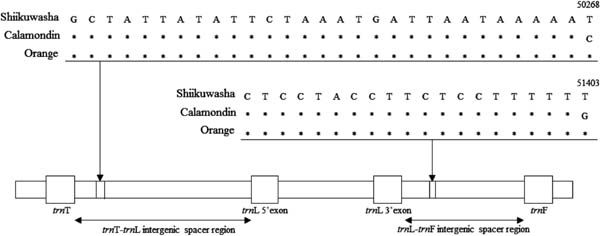
Regions corresponding to allele-specific forward primers including SNP sites in trnL-trnF and trnT-trnL of chloroplast DNA from shiikuwasha and calamondin. The numbers indicate the nucleotide positions of reference sequence (DQ864733.1) of orange. Asterisks indicate the same nucleotides among shiikuwasha, calamondin, and orange.
The trnL-trnF intergenic spacer region has been utilized as a target region for identification of several plant species using PCR-RFLP (Aragane et al., 2011; Fan et al., 2009) and PCR-SSCP (Kojoma et al., 2002). Differences in nucleotide polymorphism in the region are often found among similar plants, and species differences are utilized in plant identification (Kojoma et al., 2002). For identification of orange in juices, SNP in the trnL-trnF region was utilized with qPCR (Aldeguer et al., 2014). In addition, Taberlet et al. (1991) proposed another intergenic spacer region, trnT-trnL, and we investigated this region as well. In the present paper, sequence analysis also showed the possibility that two species-specific SNPs in the trnL-trnF and trnT-trnL regions were applicable for the identification of shiikuwasha and calamondin.
Examination of annealing temperature in allele-specific PCR To develop a method to discriminate shiikuwasha and calamondin juices, we utilized SNP sites in conserved regions of the citrus chloroplast DNA and employed PCR techniques. In the manufacturing process, juicing and heating of the product occurs, leading to fragmentation of template DNA in the juice product. Thus, the size of the amplified PCR product used to discriminate shiikuwasha and calamondin juice needed to be small. In the present paper, we adopted allele-specific PCR (Okayama et al. 1989) and designed allele-specific forward primers including the SNP site and reverse primers, leading to amplification of about 100-bp fragments (Table 2 and Fig. 2). Further, to suppress non-specific amplification, a nucleotide at the 3rd position from the 3′-end of the allele-specific forward primers was substituted with a mismatch nucleotide. The allele-specific primers were designated as CiDeLF and CiMaTL for specific detection of shiikuwasha and calamondin, respectively. To confirm the specificity of each allele-specific primer, PCR was performed using DNA prepared from shiikuwasha and calamondin peels and with various annealing temperatures for CiDeLF and CiMaTL primers from 58 to 65 °C and 54.9 to 61.2 °C, respectively. As shown in Fig. 3, by using CiDeLF primers and shiikuwasha DNA, a smaller than 100-bp fragment was found at 58–63 °C and the clearest band appeared at 61 °C. On the other hand, no amplification was confirmed by using CiDeLF primers and calamondin DNA. In the case of CiMaTL primers (Fig. 4), no amplification was confirmed using shiikuwasha DNA. However, by using calamondin DNA, fragments of about 110-bp were observed at 54.9–58.5 °C. Thus, allele-specific PCR amplification would be possible using DNA prepared from peels. Therefore, we set the annealing temperatures of amplification with CiDeLF and CiMaTL at 61 °C and 58 °C, respectively, for further experiments.
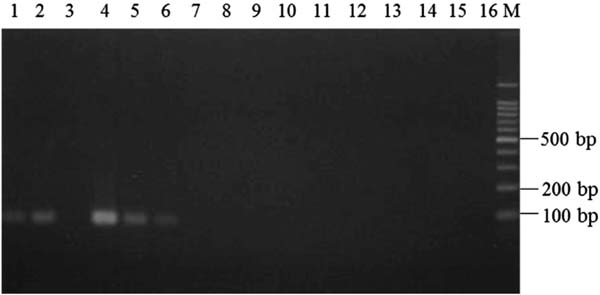
Specificity of CiDeLF primers and determination of annealing temperature for allele-specific PCR. PCR was carried out using DNA prepared from shiikuwasha (lanes 1–8) or calamondin (lanes 9–16) as the template. Lane M, 100-bp DNA ladder marker. Annealing temperature: 1, 58.0 °C; 2, 59.0 °C; 3, 60.0 °C; 4, 61.0 °C; 5, 62.0 °C; 6, 63.0 °C; 7, 64.0 °C; 8, 65.0 °C; 9, 58.0 °C; 10, 59.0 °C; 11, 60.0 °C; 12, 61.0 °C; 13, 62.0 °C; 14, 63.0 °C; 15, 64.0 °C; 16, 65.0 °C.
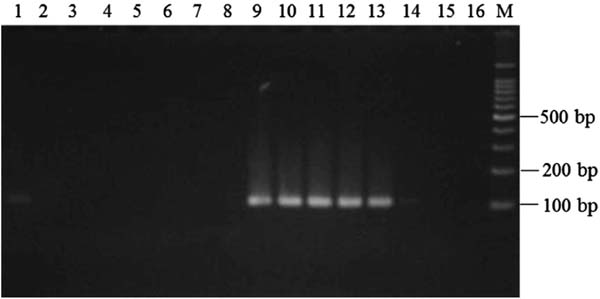
Specificity of CiMaTL primers and determination of annealing temperature for allele-specific PCR. PCR was carried out using DNA prepared from shiikuwasha (lanes 1–8) or calamondin (lanes 9–16) as the template. Lane M, 100-bp DNA ladder marker. Annealing temperature: 1, 54.9 °C; 2, 55.8 °C; 3, 56.7 °C; 4, 57.6 °C; 5, 58.5 °C; 6, 59.4 °C; 7, 60.3 °C; 8, 61.2 °C; 9, 54.9 °C; 10, 55.8 °C; 11, 56.7 °C; 12, 57.6 °C; 13, 58.5 °C; 14, 59.4 °C; 15, 60.3 °C; 16, 61.2 °C.
This allele-specific PCR technique can be performed without the need for labeled primers and qPCR machine, and can be used in SNP genotyping for species identification of organisms (Gaudet et al., 2009). Thus, the allele-specific PCR technique is expected to be a useful tool for the identification of shiikuwasha or calamondin in processed juice.
Discrimination of fruit species and hand-squeezed juice materials using allele-specific PCR We conducted allele-specific PCR for each sample using the developed allele-specific primers. Using CiDeLF primers for PCR, an approximately 80-bp product, which was estimated as 76 bp based on the sequence, was amplified using DNA from shiikuwasha peels as the template; however, it did not show the corresponding product when DNA from calamondin peels was used (Fig. 5A). In contrast, when CiMaTL primers were employed, an approx. 100-bp product was amplified using DNA from calamondin peels, but not from shiikuwasha peels (Fig. 5B). These results indicate that the designed primers resulted in specific amplification.
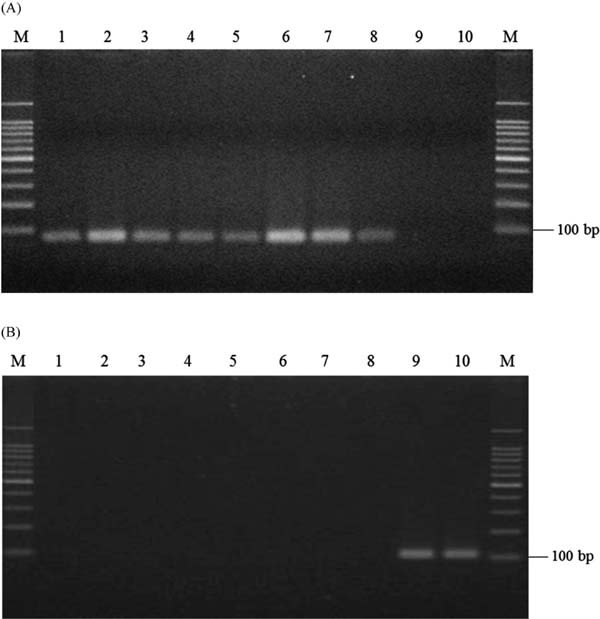
Specificity of allele-specific PCR with CiDeLF (A) or CiMaTL (B) primers using DNA prepared from citrus peels as the template. Lanes: M, 100-bp DNA ladder marker; 1, Izumi kugani; 2, Ogimi kugani; 3, Katsuyama kugani; 4, Tozanwase; 5, Kaachi; 6, B-41; 7, B-41-H; 8, shiikuwasha from Taiwan; 9, calamondin from Okinawa; 10, calamondin from Taiwan.
In application of the allele-specific PCR for discrimination of juice identity, DNA directly prepared from hand-squeezed juices was used. However, no amplification of the target regions was confirmed (data not shown). Therefore, prior to PCR, DNA from hand-squeezed juice was amplified with an Illustra GenomiPhi V2 DNA Amplification Kit and the amplified DNA was used as the template for allele-specific PCR. As shown in Fig. 6, an approx. 80-bp product was amplified by PCR using CiDeLF primers and DNA from hand-squeezed shiikuwasha juice as the template (lane 1), while no amplification was observed using DNA from calamondin as the template (lane 2). PCR using CiMaTL primers amplified an approx. 110-bp product in calamondin (lane 6), whereas no amplification was observed in shiikuwasha (lane 5).
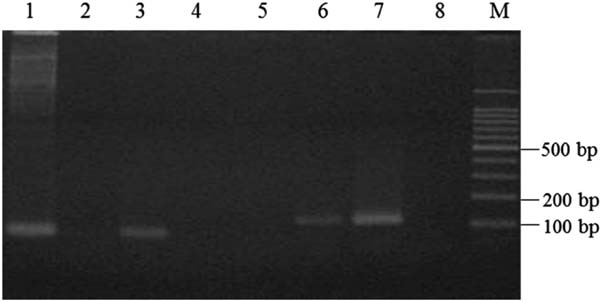
Specificity of allele-specific PCR with CiDeLF or CiMaTL primers using DNA prepared from hand-squeezed juices. Lanes: M, 100-bp DNA ladder marker; 1, hand-squeezed shiikuwasha juice; 2, hand-squeezed calamondin juice; 3, positive control (DNA from shiikuwasha); 4, negative control (without template); 5, hand-squeezed shiikuwasha juice; 6, hand-squeezed calamondin juice; 7, positive control (DNA from calamondin); 8, negative control (without template). Primers: lanes 1–4, CiDeLF; 5–8, CiMaTL.
Discrimination of materials in industrially processed and concentrated juices using allele-specific PCR DNA prepared from industrially processed and concentrated shiikuwasha and calamondin juices was also amplified with an Illustra GenomiPhi V2 DNA Amplification Kit, and the amplified DNA was used as a template for allele-specific PCR. As shown in Fig. 7, species-specific PCR products were amplified. Generally, DNA in industrially processed products such as fruit juice is thought to be partially decomposed by physical damage or chemical reaction during the manufacturing process, e.g., juicing and heating. Thus, it was difficult to amplify the target region with DNA prepared from industrially processed and concentrated juices, unlike that from citrus peel (data not shown), likely due to DNA fragmentation and/or inhibition by contaminants such as polyphenols contained in the juices (Murray et al., 1980). However, pre-amplification of DNA prepared from industrially processed and concentrated juices led to PCR amplification of species-specific target regions to discriminate citrus species in shiikuwasha and calamondin juices. These results indicate that the developed method could play a role in deterring intentional adulteration of shiikuwasha juice with calamondin juice.
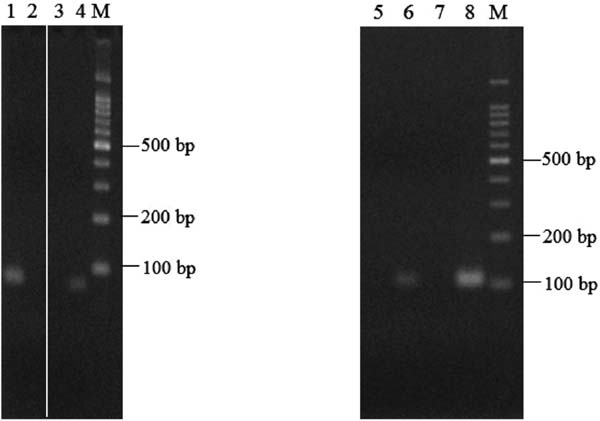
Results of allele-specific PCR with CiDeLF or CiMaTL primers using DNA prepared from industrially processed and concentrated juices. Lanes: M, 100-bp DNA ladder marker; 1, concentrated shiikuwasha juice; 2, concentrated calamondin juice from Taiwan; 3 and 7, negative control (without template); 4, positive control (DNA from shiikuwasha); 8, positive control (DNA from calamondin). Primers: lanes 1–4, CiDeLF; 5–8, CiMaTL.
PCR techniques were used to identify mandarin or orange in orange juice (Mooney et al., 2006; Ng et al., 2006). In the present study, we developed a method using DNA markers based on SNP sites on chloroplast DNA to discriminate between shiikuwasha and calamondin, even in prepared juices. Several methods utilizing chemical markers (Ogawa et al. 2001, Yamamoto et al., 2012), sensory characteristics, colorimetric values, and volatile components (Yamamoto et al., 2013) have been developed for discrimination of shiikuwasha juice adulterated with calamondin juice. Although these discrimination methods are useful techniques, in addition to these methods, development of the allele-specific PCR method based on DNA markers would strongly suppress intentional adulteration of calamondin juice to shiikuwasha juice.
In this study, in order to discriminate between shiikuwasha and calamondin fruit materials in industrially processed juice, nucleotide sequences of two intergenic spacer regions of chloroplast DNA from several types of shiikuwasha and calamondin were determined and species-specific SNP sites in both regions were identified. Allele-specific primers containing SNP sites were designed for PCR. Moreover, it was confirmed that this allele-specific PCR method can be applied not only to the citrus peel, but also to industrially processed juice with prior DNA amplification. At present, the method has been developed to discriminate only between shiikuwasha and calamondin. Although shiikuwasha juice products have not been reported to be adulterated with citrus other than calamondin, the development of other DNA markers that can be widely applied is needed to prevent future product adulteration, thereby protecting both producers and customers.
Acknowledgements This study was supported by grants from the research and development program of Promotion of Research Activities for Innovative Bioscience from Bio-oriented Technology Research Advancement Institution of Japan, the research program of Agriculture, Forestry and Fisheries of Japan.