Abstract
Over-expression of the Candida drug resistance gene CDR1 is a common mechanism generating azole-resistant Candida albicans in clinical isolates. CDR1 is transcriptionally activated through the binding of the transcription factor Tac1p to the cis-acting drug-responsive element (DRE) in its promoter. We previously demonstrated that the combination of fluconazole (FLC) and berberine (BBR) produced significant synergy when used against FLC-resistant C. albicans in vitro. In this study, we found that BBR inhibited both the up-regulation of CDR1 mRNA and the transport function of Cdr1p induced by fluphenazine (FNZ). Further, electrophoretic mobility shift assays suggested that the transcription activation complex of protein-DRE was disrupted by BBR, and electrospray ionization mass spectrometry analysis showed that BBR bound to the DRE of CDR1. Thus we propose that BBR inhibits the FNZ-induced transcriptional activation of CDR1 in C. albicans by blocking transcription factor binding to the DRE of CDR1. These results contribute to our understanding of the mechanism of synergistic effect of BBR and FLC.
Candida albicans is one of the major opportunistic fungal pathogens in humans, and causes candidiasis ranging from superficial mucosal infections to life-threatening systemic disorders.1–3) Triazole antifungal agents, such as fluconazole (FLC) and itraconazole, are considered to be the first-choice agents for treatment and prophylaxis because of their relatively low side effects and high effectiveness on mucosal infections. However, with prolonged exposure to azoles, drug resistance becomes a serious problem in clinical practice.4,5) Several genes have been confirmed to contribute to the development of multidrug resistance including ERG11, CDR1, CDR2, MDR1, RTA2, AOX1 and AOX2.6–12) Among them, up-regulation of CDR1, a member of the ATP-binding cassette (ABC) transporter super-family in C. albicans, is a common occurrence in clinical C. albicans isolates resistant to antifungal azoles.13) Cdr1p pumps azoles out of C. albicans cells to reduce intracellular accumulation, which can be indirectly identified by the efflux of rhodamine 6G (R6G), which is a fluorescent substrate transported by Cdr1p.14) In the promoter of CDR1, the basal expression element (BEE) (−960–−710) is responsible for basal transcription of CDR1, and the drug-responsive element (DRE) (−264–−244) is responsible for drug-induced transcription.15) The DRE (5′-CGG ATA TCG GAT ATT TTT TTT-3′) contains CGG triplets that are often recognized by Zinc finger proteins.16,17) It has been demonstrated that the DRE of CDR1 can be recognized and bound by the transcription controller Tac1p that is characterized by a highly conserved Zn(II)2Cys6 motif within the DNA binding domain (DBD).15,16)
Small molecules which can regulate the expression of CDR1 become a promising strategy to combat azole resistance in C. albicans. Berberine (BBR) is a plant-derived isoquinoline alkaloid from the roots and bark of medicinal plants such as Rhizoma coptidis and Phellodendron amurense. Although its traditional use mainly focused in gastroenteritis and secretory diarrhea, antifungal activity with low cytotoxicity to human cells has recently been reported.18–20) The use of antimicrobial agents in combination has been a common practice to improve therapeutic efficacy, reduce toxic side-effects of these drugs, and decrease drug resistance. A synergistic antifungal effect has been confirmed for the combination of BBR and amphotericin B in an infectious mouse model.21) In vitro, synergism between BBR and miconazole has been reported against both planktonic and biofilm C. albicans.22) Moreover, the combination of FLC and BBR produced significantly synergistic effect against both FLC-sensitive and FLC-resistant C. albicans in vitro.23,24) However, the molecular target of BBR remains unknown.
As BBR has been reported as a DNA binding molecule, its mechanism of action has been correlated to the biophysical parameters of DNA binding.25–28) In this study, we investigated whether BBR regulated the transcriptional expression of CDR1 by binding to the promoter of CDR1. We first explored the inhibitory effect of BBR on the transcriptional activity of CDR1 and CDR2. The influence by BBR of the binding of protein extracts to the DRE of CDR1 was characterized through an electrophoretic mobility shift assay (EMSA), and the binding of BBR to the CDR1 DRE was further confirmed through electrospray ionization mass spectrometry (ESI-MS) analysis.
MATERIALS AND METHODS
Antifungal AgentsBBR and fluphenazine (FNZ) were obtained from Sigma (St. Louis, MO, U.S.A.). Stock solutions were prepared in dimethyl sulfoxide, then filter sterilized and stored at −70°C.
Strains and MediaC. albicans strain SC5314 was kindly provided by William A. Fonzi (Department of Microbiology and Immunology, Georgetown University, Washington, D.C., U.S.A.). C. albicans strains were grown in liquid YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C with shaking (200 rpm).
Relative Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR) (qPCR)A single colony of C. albicans SC5314 was grown in YPD medium in the presence or the absence of 1 µM or 10 µM BBR at 30°C with shaking overnight for 16 h. Late-logarithmic phase cells were then treated with or without 50 µg/mL of FNZ for 30 min to induce the expression of CDR1 and CDR2.29) All the primer sequences are listed in Table 1. Total RNA isolation, purification, and PCR amplification were performed as described previously.30) The 18S ribosomal RNA (rRNA) was used as an internal control, and the relative fold change of target genes in the strains with the treatment of BBR, or FNZ, or both relative to that of the genes in the strains without any treatment was expressed as 2−ΔΔCT. Triplicate independent experiments were performed using a 7500 Real Time PCR system (Applied Biosystems) and SYBR Green I (TaKaRa Bio, Tokyo, Japan).
Table 1. Primers Sequences Used in This Study
| Primer name | Sequence | Amplicon size (bp) |
|---|
| 18S rRNA-F | TCTTTCTTGATTTTGTGGGTGG | 150 |
| 18S rRNA-R | TCGATAGTCCCTCTAAGAAGTG | |
| CDR1-F | GATTCTCAA ACTGCCTGGTC | 116 |
| CDR1-R | CCAAAATAAGCCGTTCTTCCAC | |
| CDR2-F | ACTGCAAGTCACAACATCC | 132 |
| CDR2-R | GAATGGACTAACACCAGGTATG | |
The glucose-induced efflux of R6G (Sigma) from C. albicans strains was investigated as described previously31) with some modifications. A single colony of C. albicans SC5314 was grown in YPD medium in the presence or the absence of 1 µM or 10 µM BBR at 30°C with shaking for 16 h. The cells were collected, washed with phosphate-buffered saline (PBS) buffer, resuspended in PBS buffer to the concentration of 1.5×107 cells/mL, and cultured for 2 h to deplete ATP stores. R6G was added to a final concentration of 10 µM and the suspensions were then incubated for another 2 h to allow R6G accumulation under glucose starvation conditions. C. albicans cells were then treated with or without 50 µg/mL of FNZ for 30 min. The starved cells were washed twice in PBS buffer, and portions (1 mL) were incubated at 30°C for 5 min before the addition of 2 mM glucose to initiate R6G efflux. At specified intervals after the addition of glucose, the cells were removed by centrifugation. Triplicate 100 µL of the cell supernatants were transferred to 96-well flat-bottom microtiter plates on a Polarstar Optima instrument (BMG Labtechnologies, Offenburg, Germany). The fluorescence densities of three independent samples were measured with excitation wavelength at 515 nm and an emission wavelength at 555 nm using a Polarstar Optima instrument (BMG Labtech, Offenburg, Germany). The concentration of R6G was calculated using a standard concentration curve.
EMSAA single colony of C. albicans SC5314 was grown in liquid YPD medium to reach 1.5×107 cells/mL. Cells were treated with or without 50 µg/mL of FNZ for 30 min. The total proteins were extracted and quantified as described previously.15) The oligonucleotide 5′-TTG AGA CGG ATA TCG GAT ATT TTT TTG-3′ containing the DRE of CDR1 was end-labeled with digoxin (DIG) and then annealed to its complement to produce the oligonucleotide probe. A total of 10 µM probe was preincubated with 10 µM, 100 µM or 1 mM BBR (molecular weight (MW) 336) in EMSA binding-buffer (Beyotime, Jiangsu, China) for 1.5 h at 0°C. A total of 40 µg protein extracts from cells with or without the treatment of FNZ were added to the buffer. After incubation for additional 1.5 h at 0°C, the mixtures were loaded onto a non-denaturing 6% polyacrylamide gel (PAGE). The gel was then run in 0.5×TBE buffer at 4°C for 1 h and the separated proteins transferred to a positively charged nylon membrane. The oligo probes were detected with DIG Nucleic acid Detection Kit (Innogent, Shenzhen, China) according to the manufacturer’s manual.
ESI-MS AnalysisSingle strand oligonucleotides were synthesized, annealed to their complements, followed by purification with PAGE. A stock solution of 100 µM double-strand DNAs (dsDNAs) (Table 2) was then produced (Biosune, Shanghai, China). A mixture of 25 µL of stock solution and 5 µL of 2 mM BBR was added to a solution of methanol-100 mM aqueous ammonium acetate (20 : 80, v/v) and incubated for 30 min at 25°C. In the controls, the BBR solution was replaced by dimethyl sulfoxide.
Table 2. DsDNAs Used in This Study
| Name | Sequence | Position in CDR1 promoter | GC content | MW (Da) |
|---|
| dsDNA1 | 5′-GACGGATATCGGAT-3′/ 5′-ATCCGATATCCGTC-3′ | −458–−445 | 50.0% | 8526.7 |
| dsDNA2 | 5′-GATATTTTTTTG-3′/ 5′-CAAAAAAATATC-3′ | −446–−435 | 16.7% | 7287.0 |
ESI-MS spectra were obtained using a Finnigan LCQDeca XP Max ion trapmass spectrometer (Thermo Finnigan, San Jose, CA, U.S.A.). The samples were directly infused into the mass spectrometer at a flow rate of 3 µL/min. All the experiments were performed in negative ion mode. The spray voltage was set to 3.5 kV and the capillary temperature was set at 220°C. Nitrogen gas was used to ensure efficient desolvation. The scanned range was from 900 to 2000 m/z, and helium (40 arb) was introduced as the trapping gas. Data were collected and analyzed with the Xcalibur software (Thermo Finnigan).
RESULTS
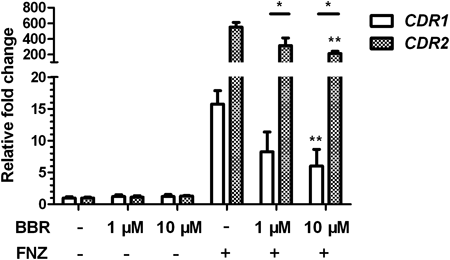
BBR Inhibits the FNZ-Induced Up-Regulation of CDR1 and CDR2 mRNACdr1p and Cdr2p are multidrug transporters of the ABC super-family in C. albicans. CDR1 shows a basic expression level under normal conditions, while CDR2 transcript is not detectable. Both genes can be rapidly transcriptionally induced by FNZ.15) Relative qPCR reactions were performed to investigate whether BBR could influence the expression of CDR1 or CDR2. As shown in Fig. 1, there were no obvious up- or down-regulation of either CDR1 or CDR2 in the strains incubated with or without BBR alone for 16 h. However, after the treatment of FNZ, the mRNA level of CDR1 was increased 15.7 fold, while preincubation with 1 µM or 10 µM BBR for 16 h reduced the FNZ up-regulated mRNA levels of CDR1 by 47.6% and 61.8% to 8.23 and 6.01 fold, respectively. Conspicuously, the mRNA level of CDR2 was induced 550 fold with the treatment of FNZ, whereas preincubation with BBR significantly reduced the FNZ-induced CDR2 mRNA by 43% and 61.5%. As the minimal inhibitory concentration (MIC80) of BBR alone against C. albicans reaches 64 µg/mL (190 µM), while that of BBR combined with FLC is 1 µg/mL (3 µM), the concentrations of BBR used here were low. These results suggested that BBR with low concentrations (1 µM or 10 µM) did not affect the basic transcription of CDR1 or CDR2, but inhibited their FNZ-induced up-regulation.
R6G is a fluorescent substrate of Cdr1p and its efflux is positively correlated with the expression level of Cdr1p.14) Since BBR inhibited the up-regulation of FNZ-induced CDR1 mRNA, we investigated if BBR influenced the transport function of Cdr1p as measured through R6G efflux. Efflux from de-energized R6G-preloaded cells required the presence of glucose. Efflux of R6G from the strains with the treatment of BBR alone increased less than 1 fold in 20 min and then decreased within 70 min (data not shown), similar to that from strains without any treatment (Fig. 2; none group). However, by 50 min following glucose addition the extracellular R6G concentration increased significantly with the treatment of FNZ alone (p<0.01; Fig. 2). In contrast, the degree of FNZ-induced efflux of R6G in strains preincubated with 1 µM and 10 µM BBR was lower than that in strains without preincubation (p<0.05; Fig. 2). These results provided evidence that BBR inhibited energy-dependent extrusion of their substrates, probably resulting from the decreased FNZ-induced up-regulation of CDR1.

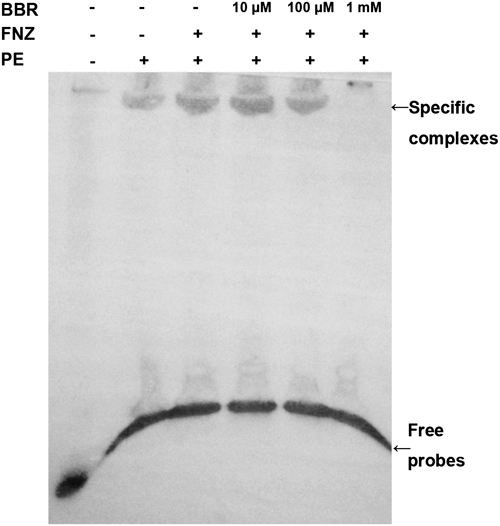
Previous studies had demonstrated that the Tac1-binding DRE site in the CDR1 promoter consisted of a direct CGG repeat with four intervening nucleotides (CGG AAA TCG G).16) To explore whether BBR affects DRE function, an EMSA assay was performed using a 27 bp DIG-labeled probe containing the DRE of CDR1. In order to get clear bands, the concentration of both the DIG-labeled probe and BBR needed to be high. As shown in Fig. 3, bands of specific complexes were obtained with protein extracts from both FNZ-treated and -untreated strains. The identified complex was of a slightly higher intensity with extracts from a FNZ-treated strain. However, a preincubation of 100 µM BBR with the probe resulted in a slightly reduced band intensity when compared those assays without BBR or with a 10 µM BBR incubation. Furthermore, the band was greatly reduced when the preincubation concentration of BBR reached 1 mM. The edge zone effects were observed in bands of free probes. These results suggested that BBR prevented, in a dose-dependent manner, proteins from binding to the DRE in the CDR1 promoter, leading to the disruption of transcriptional activation of CDR1 by BBR.
ESI-MS is a powerful tool to measure small molecules binding to specific dsDNA sequences.32) We divided the DRE sequence found in the promoter of CDR1 into two parts: dsDNA1 of a 14 bp sequence containing CGG triplets in direct repeats and dsDNA2 consisting of 12 bp sequence rich in AT bases. Both dsDNAs were used to monitor binding of BBR to the DRE by negative ion ESI-MS. In each spectrum, the relative abundances of the relevant ions have been normalized to 100% for the generally most abundant ion. The ESI-MS spectra of the free dsDNA1 and dsDNA2 were recorded. Three main ions at m/z 1399, 1442 and 1704 in dsDNA1 spectrum (Fig. 4A) and two main ions at m/z 1217 and 1826 in dsDNA2 spectrum (Fig. 4B) were predominant in the scanned range (m/z 900–2000). The ions at m/z 1704 and 1826 can be easily assigned as ions of dsDNA1 with 5 negative charges ([dsDNA15−]) and dsDNA2 with 4 negative charges ([dsDNA24−]), compared to ions corresponding to the single-stranded (ss) oligonucleotides (m/z 1399 ss3−, 1442 ss3− and 1217 ss3−). The dsDNA ions indicated the formation of DNA duplexes in the annealing process. When the dsDNAs were mixed with BBR at molar ratios 1 : 4 and incubated for 30 min at 25°C, the binding spectra showed ions at m/z 1772 in the dsDNA1 binding spectrum (Fig. 4C), identified as [dsDNA1+BBR]5−, and ions at m/z 1904 in the dsDNA2 binding spectrum (Fig. 4D), identified as [dsDNA2+BBR]4−. The two ions of [dsDNA1+BBR]5− and [dsDNA2+BBR]4− revealed not only the existence of the BBR-dsDNA binding complexes, but also the low strands specificity of BBR binding to dsDNA. These results suggested that the DNA binding effect of BBR might result in the disruption of transcriptional activity by preventing the transcription factors such as Tac1p from binding to the DRE of CDR1.

DISCUSSION
Several studies have reported that BBR, together with azoles or amphotericin B, exerts antifungal activity against C. albicans.21–24) We previously revealed that BBR in combination with FLC augmented the endogenous reactive oxygen species (ROS) production through enhancing the tricarboxylic acid cycle and inhibiting ATP-synthase activity.33) Nevertheless, in the search for the mechanism of synergism between BBR and antifungal agents, their individual antifungal activity needs to be taken into consideration. The antimicrobial mechanism of BBR has been reported to involve binding to DNA by partial interaction with AT-rich sequences.25) In this study, we have provided evidence that BBR blocked the binding of transcriptional factors to the DRE in the promoter of CDR1. Instead, BBR associated with the DRE of CDR1 and disrupted the transcriptional activity and efflux function of Cdr1p.
The reduced intracellular accumulation of drugs is a common mechanism of drug resistance, which is correlated with the increased expression of CDR1 and CDR2. Tac1p is the major transcription factor needed for the up-regulation of CDR1 and CDR2.15,16) Thus it was expected that BBR inhibited the up-regulation of FNZ-induced CDR1 and CDR2, but did not affect the basic transcription of CDR1 or CDR2. The combining of BBR with DRE of CDR1 did not influence the basic expression, but prevented transcriptional factors, possibly Tac1p, from further binding to the DRE. This might also reveal a mechanism of synergistic effect of BBR and FLC.
We previously have explored the small molecule approach for gene regulation to overcome drug resistance in C. albicans. A cell-permeable, sequence-specific, DNA-binding polyamide SL-A92 was designed and synthesized to target the sequence CGG, which is the binding site of zinc cluster transcription factors.34) SL-A92 decreased the transcriptional activity of CDR1 and thus blocked the development of drug resistance in the C. albicans strain SC5314. Nevertheless, SL-A92 had neither antifungal activity nor effects on strain growth and did not inhibit the drug resistance of a clinical isolate. Compared to polyamide SL-A92, BBR, identified as a DNA-binder in C. albicans, has individual and synergistic antifungal effects, which open further avenues for exploring the potential therapeutic application of BBR.
In summary, BBR’s binding to the DRE probably resulted in the blockage of the transcriptional activation of CDR1, thus leading to the inhibition of up-regulation of CDR1. However, further study is needed to explore the possible interactions between BBR and the transcriptional factors. This will be helpful to develop BBR into a potential agent against drug resistance in C. albicans.
Acknowledgment
We are grateful to William A. Fonzi for kindly providing the C. albicans strain SC5314. We also thank Malcolm Whiteway (Concordia University) for critical reading. This work was supported by the Grants from the China National 973 Program (2013CB531602), National Natural Sciences Fund of China (No. 30772644, 21072226, 21272270, 31000079), Shanghai Pujiang Program (No. 05PJ14003).
REFERENCES
- 1) Garber G. An overview of fungal infections. Drugs, 61 (Suppl. 1), 1–12 (2001).
- 2) Maertens J, Vrebos M, Boogaerts M. Assessing risk factors for systemic fungal infections. Eur. J. Cancer Care (Engl.), 10, 56–62 (2001).
- 3) Richardson MD. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother., 56 (Suppl. 1), i5–i11 (2005).
- 4) Boken DJ, Swindells S, Rinaldi MG. Fluconazole-resistant Candida albicans. Clin. Infect. Dis., 17, 1018–1021 (1993).
- 5) White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev., 11, 382–402 (1998).
- 6) Du W, Coaker M, Sobel JD, Akins RA. Shuttle vectors for Candida albicans. control of plasmid copy number and elevated expression of cloned genes. Curr. Genet., 45, 390–398 (2004).
- 7) Jia XM, Ma ZP, Jia Y, Gao PH, Zhang JD, Wang Y, Xu Y, Wang L, Cao YY, Cao YB, Zhang LX, Jiang YY. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem. Biophys. Res. Commun., 373, 631–636 (2008).
- 8) Jia XM, Wang Y, Jia Y, Gao PH, Xu YG, Wang L, Cao YY, Cao YB, Zhang LX, Jiang YY. RTA2 is involved in calcineurin-mediated azole resistance and sphingoid long-chain base release in Candida albicans. Cell. Mol. Life Sci., 66, 122–134 (2009).
- 9) Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet., 27, 320–329 (1995).
- 10) Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology, 143, 405–416 (1997).
- 11) Wirsching S, Michel S, Morschhauser J. Targeted gene disruption in Candida albicans wild-type strains. the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol., 36, 856–865 (2000).
- 12) Yan L, Li M, Cao Y, Gao P, Cao Y, Wang Y, Jiang Y. The alternative oxidase of Candida albicans causes reduced fluconazole susceptibility. J. Antimicrob. Chemother., 64, 764–773 (2009).
- 13) Krishnamurthy S, Gupta V, Snehlata P, Prasad R. Characterisation of human steroid hormone transport mediated by Cdr1p, a multidrug transporter of Candida albicans, belonging to the ATP binding cassette super family. FEMS Microbiol. Lett., 158, 69–74 (1998).
- 14) Maesaki S, Marichal P, Vanden Bossche H, Sanglard D, Kohno S. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother., 44, 27–31 (1999).
- 15) de Micheli M, Bille J, Schueller C, Sanglard D. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol., 43, 1197–1214 (2002).
- 16) Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell, 3, 1639–1652 (2004).
- 17) MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators. the zinc cluster proteins. Microbiol. Mol. Biol. Rev., 70, 583–604 (2006).
- 18) Mantena SK, Sharma SD, Katiyar SK. Berberine, a natural product, induces G1-phase cell cycle arrest and caspase-3-dependent apoptosis in human prostate carcinoma cells. Mol. Cancer Ther., 5, 296–308 (2006).
- 19) Ficker CE, Arnason JT, Vindas PS, Alvarez LP, Akpagana K, Gbeassor M, De Souza C, Smith ML. Inhibition of human pathogenic fungi by ethnobotanically selected plant extracts. Mycoses, 46, 29–37 (2003).
- 20) Imanshahidi M, Hosseinzadeh H. Pharmacological and therapeutic effects of Berberis vulgaris and its active constituent, berberine. Phytother. Res., 22, 999–1012 (2008).
- 21) Han Y, Lee JH. Berberine synergy with amphotericin B against disseminated candidiasis in mice. Biol. Pharm. Bull., 28, 541–544 (2005).
- 22) Wei G-X, Xu X, Wu CD. In vitro synergism between berberine and miconazole against planktonic and biofilm Candida cultures. Arch. Oral Biol., 56, 565–572 (2011).
- 23) Iwazaki RS, Endo EH, Ueda-Nakamura T, Nakamura CV, Garcia LB, Filho BP. In vitro antifungal activity of the berberine and its synergism with fluconazole. Antonie van Leeuwenhoek, 97, 201–205 (2010).
- 24) Quan H, Cao Y-Y, Xu Z, Zhao J-X, Gao P-H, Qin X-F, Jiang Y-Y. Potent in vitro synergism of fluconazole and berberine chloride against clinical isolates of Candida albicans resistant to fluconazole. Antimicrob. Agents Chemother., 50, 1096–1099 (2006).
- 25) Davidson MW, Lopp I, Alexander S, Wilson WD. The interaction of plant alkaloids with DNA. II. Berberinium chloride. Nucleic Acids Res., 4, 2697–2712 (1977).
- 26) Debnath D, Kumar GS, Nandi R, Maiti M. Interaction of berberine chloride with deoxyribonucleic acids: evidence for base and sequence specificity. Indian J. Biochem. Biophys., 26, 201–208 (1989).
- 27) Saran A, Srivastava S, Coutinho E, Maiti M. 1H NMR investigation of the interaction of berberine and sanguinarine with DNA. Indian J. Biochem. Biophys., 32, 74–77 (1995).
- 28) Bhadra K, Maiti M, Kumar GS. Berberine-DNA complexation: New insights into the cooperative binding and energetic aspects. Biochim. Biophys. Acta, 1780, 1054–1061 (2008).
- 29) Saidane S, Weber S, De Deken X, St-Germain G, Raymond M. PDR16-mediated azole resistance in Candida albicans. Mol. Microbiol., 60, 1546–1562 (2006).
- 30) Yan L, Cote P, Li XX, Jiang YY, Whiteway M. PalI domain proteins of Saccharomyces cerevisiae and Candida albicans. Microbiol. Res., 167, 422–432 (2012).
- 31) Yan L, Zhang J, Li M, Cao Y, Xu Z, Cao Y, Gao P, Wang Y, Jiang Y. DNA microarray analysis of fluconazole resistance in a laboratory Candida albicans strain. Acta Biochim. Biophys. Sin. (Shanghai), 40, 1048–1060 (2008).
- 32) Wang Z, Guo X, Liu Z, Cui M, Song F, Liu S. Studies on alkaloids binding to GC-rich human survivin promoter DNA using positive and negative ion electrospray ionization mass spectrometry. J. Mass Spectrom., 43, 327–335 (2008).
- 33) Xu Y, Wang Y, Yan L, Liang RM, Dai BD, Tang RJ, Gao PH, Jiang YY. Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J. Proteome Res., 8, 5296–5304 (2009).
- 34) Zhu SL, Jiang ZH, Gao PH, Qiu Y, Wang L, Jiang YH, Zhang DZ. A novel polyamide SL-A92 as a potential fungal resistance blocker: synthesis and bioactivities in Candida albicans. Acta Pharmacol. Sin., 31, 855–860 (2010).




