2016 年 39 巻 1 号 p. 25-32
2016 年 39 巻 1 号 p. 25-32
Gene targeting via homologous recombination, albeit highly inefficient in human cells, is considered a powerful tool for analyzing gene functions. Despite recent progress in the application of artificial nucleases for genome editing, safety issues remain a concern, particularly when genetic modification is used for therapeutic purposes. Therefore, the development of gene-targeting vectors is necessary for safe and sophisticated genetic modification. In this paper, we describe the effect of vector structure on random integration, which is a major obstacle in efficient gene targeting. In addition, we focus on the features of exon-trapping-type gene-targeting vectors, and discuss a novel strategy for negative selection to enhance gene targeting in human cells.
Gene targeting is a technique that utilizes homologous recombination to modify endogenous gene loci by introducing targeting vectors into a cell.1) This technology is useful for analyzing gene functions and may potentially be applied to human gene therapy.2) However, the efficiency of gene targeting is extremely low in human cells primarily due to the low frequency of targeted integration. Additionally, the high frequency of random integration, which involves the random insertion of a transfected vector into chromosomal DNA, poses a challenge to gene targeting in human cells (Fig. 1). Recently, the development of artificial nucleases such as transcription activator-like effector nuclease (TALEN) and clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) has dramatically improved genetic modification technologies in cells derived from various species.3,4) These nucleases are designed to specifically induce a DNA double-strand break (DSB) at the desired site of a target gene; however, the use of artificial nucleases, albeit highly efficient, has been shown to be associated with off-target mutations,5–8) which lead to altered expression or function of non-target genes, thereby giving rise to safety concerns. Importantly, the use of artificial nucleases is compatible with conventional gene-targeting methods, since DSB induction in the target gene can also enhance gene targeting when a targeting vector is co-transfected into the cells. Therefore, it is valuable and necessary to develop sophisticated targeting vectors for safe or complex genetic modifications. In this paper, we outline methods for constructing targeting vectors for efficient genetic modification, with an emphasis on exon-trap vectors and a novel negative selection strategy to enhance gene targeting in human cells.
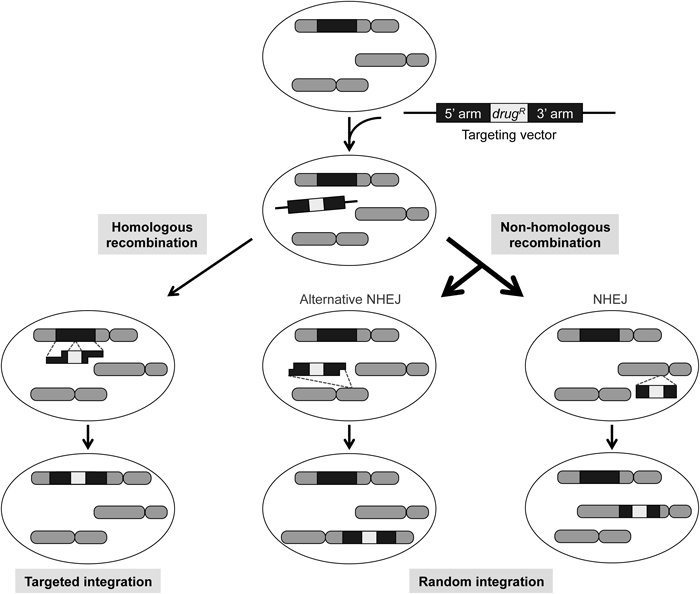
When a targeting vector is transfected into human cells, random integration by non-homologous recombination occurs at least 2 to 3 orders of magnitude more frequently than the targeted integration via homologous recombination. Thus, random integration is a major obstacle to efficient gene targeting. The non-homologous end-joining (NHEJ) pathway has been considered responsible for random integration; however, recent evidence indicates that an additional mechanism, termed alternative NHEJ, significantly contributes to this process. For homologous recombination to occur, extensive 5′-end resection is a prerequisite for producing 3′-single stranded DNA (ssDNA) overhangs that act to invade into a homologous sequence in the genome. Mechanistically, the alternative NHEJ pathway typically accompanies 5′-end resection (albeit limited), while NHEJ does not (note that ssDNA regions produced in the homology arms are drawn shorter for alternative NHEJ than for homologous recombination). drugR, drug-resistance gene.
The efficiency of gene targeting is calculated by dividing targeted integration frequency by the sum of random integration frequency and targeted integration frequency (targeted integration / (random integration+targeted integration)). Since random integration occurs at an extremely high frequency in human cells,9) controlling both the recombination mechanism involved in random integration or the occurrence of random integrants is an effective strategy to improve the efficiency of gene targeting. Random integration has been thought to occur via non-homologous end joining (NHEJ), which is a major DSB repair pathway.10,11) A targeting efficiency of nearly 100% has been reported in lower eukaryotes (such as Neurospora crassa) lacking NHEJ.12,13) We were able to increase the targeting efficiency in human cells lacking NHEJ; however, the frequency of random integration in these NHEJ-deficient cells failed to decrease,14) suggesting that alternative mechanisms are involved in NHEJ-independent random integration.15,16) Elucidation of the mechanisms underlying NHEJ-dependent and -independent random integration is important for increasing the efficiency of gene targeting in human cells.
In general, targeting vectors carry a positive selection marker (e.g., a drug-resistance gene) placed between homologous DNA sequences in the 5′-upstream and 3′-downstream region of the target locus (5′ arm and 3′ arm)17) (Fig. 2). An additional method that is a widely used for efficiently selecting homologous recombinants involves inserting a suicide gene (or a drug-sensitive gene) outside the homologous region.18) Commonly used vectors include plasmid vectors and viral vectors such as the adeno-associated virus. In general, plasmid vectors are safe and versatile compared with viral vectors. In this section, we describe the features of, and challenges associated with, plasmid-based targeting vectors, and focus on how vector structure affects integration frequencies.
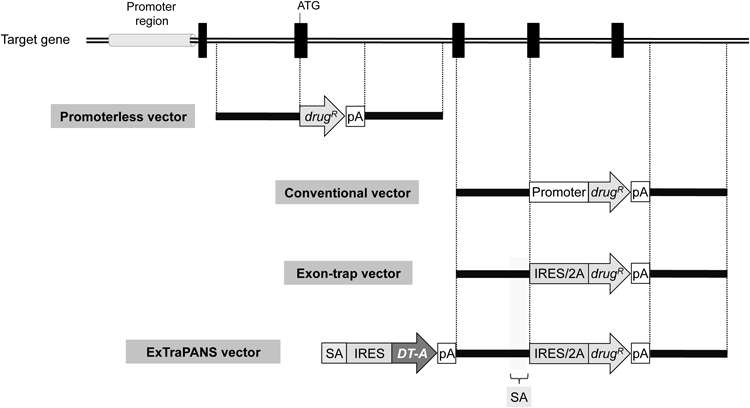
Conventional gene-targeting vectors carry a positive selection marker (i.e., drug-resistance gene, drugR), with a promoter and a polyA sequence (pA), between the 5′ and 3′ homology arms. These vectors also contain a negative selection marker outside the arm(s) (not shown in this figure; see the text for details). Promoterless vectors are different from conventional vectors in that the drug-resistance gene does not have a promoter. The design of promoterless vectors requires that the positive selection marker is inserted in-frame to a coding sequence, usually at the initiation codon of the target gene (shown as “ATG”). Exon-trap vectors also do not have a promoter attached to the drug-resistance gene. These vectors instead carry a splice acceptor site (SA) to trap the splicing from an upstreamly located splice donor site, along with an IRES or 2A peptide sequence. More specifically, the 5′ arm contains a sequence that serves as an SA site directly upstream of the drug-resistance gene. The IRES sequence allows for a cap-independent translation of mRNA, whereas the 2A peptide sequence encodes a conserved motif (DV/IEXNPG*P, in which * indicates where a peptide bond is not formed) that enables an efficient and equimolar synthesis of two different proteins from a single open reading frame. ExTraPANS vectors additionally consist of a gene cassette upstream of the 5′ arm of exon-trap vectors. This cassette is composed of an SA site, an IRES sequence, a DT-A gene and a polyA sequence, and serves as a negative selection marker. When an ExTraPANS vector integrates non-homologously into a gene-coding region, the upstream SA site traps the splicing from an upstream exon to allow DT-A expression, thereby killing random integrants. A gray cylinder and black boxes represent promoter region and exons, respectively.
The efficiency of gene targeting is directly proportional to the length of the homology arms, such that longer arms are generally associated with more efficient gene targeting. A positive correlation between arm length and targeting efficiency has previously been reported in mouse ES cells.1) A similar trend has also been reported in lower eukaryotes such as Saccharomyces cerevisiae and N. crassa.13,19) However, this correlation has not yet been fully examined in human cells due to the extremely low frequency of targeted integration. Recently, we examined the relationship between the length of homology arms and targeting efficiency using a human pre-B lymphoma cell line, Nalm-6.20) Our data showed that the frequency of targeted integration increased with increase in arm length, but so did the frequency of random integration.20) Therefore, shorter arms may be preferable for suppressing random integration; however, in some cases, there is no other option but to design a vector with long arms. The vector design strategy is discussed in detail in Section 4.
As stated above, the frequency of random integration is considerably high even in human cells lacking NHEJ.14) In order to elucidate the mechanism underlying this phenomenon, we used vectors with a variety of arm lengths and compared the frequencies of random integration between these vectors. The data showed that random integration occurs more frequently when vectors with longer arms are used, particularly when the vectors contain large amounts of repetitive DNA sequences (which include, but are not limited to, short interspersed nucleotide elements (SINEs) and long interspersed nucleotide elements (LINEs)).20) This finding suggests that the presence of repetitive DNA sequences may promote random integration, presumably via alternative NHEJ (alt-NHEJ), which is an alternative DSB repair pathway. As the alt-NHEJ pathway typically involves 5′-end resection of a DSB (see Fig. 1) and is believed to favor microhomology-mediated joining of resected DNA ends,21,22) it is possible that microhomologies between repetitive DNA sequences of the vector and the target genome enhance alt-NHEJ-mediated random integration. Although the detailed molecular mechanisms of this reaction are unclear, experiments using targeting vectors containing no repetitive sequences or those comprising repetitive sequences alone may be useful in developing vectors that do not or rarely exhibit random integration.
3.2 Principles of, and Challenges Associated with, Positive and Negative SelectionIn general, targeting vectors carry a positive selection marker, such as a drug-resistance gene (e.g., puromycin-resistance gene, hygromycin-resistance gene, or neomycin-resistance gene), attached to a promoter. As a result, most vectors inserted into the genome confer resistance to a particular antibiotic, according to the selection marker utilized. To counterselect random integrants, a strategy termed negative selection is preferentially employed, in which a suicide gene such as a diphtheria toxin A fragment gene (DT-A) or a drug-sensitive gene such as a herpes simplex virus thymidine kinase gene (HSV-tk) is added to the vector outside the arm(s).18,23) In particular, the use of DT-A is advantageous in that negative selection utilizing this gene does not require the presence of a drug. However, the occurrence of strong cytotoxicity after transfection, presumably due to transient DT-A expression before vector integration, poses a challenge. In addition, this negative selection strategy becomes ineffective when gene silencing occurs after vector integration. Therefore, the effects of DT-A on gene targeting (i.e., on selecting homologous recombinants) remain unclear.
3.3 Characteristics and Effectiveness of Vectors with a Promoterless Marker GeneMethods that reduce the frequency of random integration include the use of promoterless vectors and exon-trapping-type targeting vectors (exon-trap vectors), which both rely on a positive selection marker that lacks a promoter24–26) (Fig. 2). When these vectors are inserted into a genomic region with no or low transcriptional activity, cells do not acquire drug resistance. As a result, a single promoterless gene serves as a marker for both positive and negative selection. However, the construction of promoterless vectors requires considerable time and effort, as these vectors need to be designed to enable the positive selection marker to be inserted in frame to an exon of the target gene (Fig. 2).
Similar to promoterless vectors, exon-trap vectors do not have their own promoter, but possess a splice acceptor (SA) site as well as an internal ribosome entry site (IRES) sequence27) or a 2A peptide sequence28–30) attached upstream of the positive selection marker, thus enabling polycistronic expression (i.e., expression of the marker gene) in a manner dependent on promoter activity of the target gene (Fig. 2). With regards to IRES sequences, the reading frames of the target gene and the drug-resistance gene do not need to match, rendering vector construction relatively easy. In contrast, these reading frames are required to match when a 2A peptide sequence is employed. Therefore, the construction of 2A peptide-based vectors requires more effort than the construction of IRES-based vectors. However, the use of 2A peptide sequences is advantageous over IRES sequences in that these sequences are expected to provide more efficient marker gene expression for target genes with low expression levels.31)
In order to investigate the effectiveness of exon-trapping gene targeting, we performed gene-targeting experiments in a variety of human cell types. In previous studies, targeting efficiency using conventional vectors was shown to be approximately 0.2% in HT1080 cells32) and less than 0.1% in induced pluripotent stem (iPS) cells.33) In contrast, we found that the targeting efficiency in those cells was ca. 1–5% when exon-trap vectors were employed, by virtue of reduced random-integration frequencies34) (Table 1). In human Nalm-6 cells, targeting efficiency was increased to ca. 25–100% with the use of exon-trap vectors (unpublished data). Intriguingly, such ultrahigh-efficiency gene targeting associated with exon-trap vectors has also been reported in mouse ES cells (>50% on average).24) Collectively, these data unequivocally indicate that exon-trap vectors provide a useful and versatile tool for efficient gene targeting.
| Human cell type | Conventional vector | Exon-trap vector | ExTraPANS vector |
|---|---|---|---|
| HT1080 | 6.6 | 1 | 0.06 |
| HeLa | NT (>10)* | 1 | 0.10 |
| Nalm-6 | 6.4 | 1 | 0.52 |
| iPS | 4.4 | 1 | 0.24 |
Targeting vectors for the human HPRT gene were each transfected into HT1080 cells, HeLa cells, Nalm-6 cells, or iPS cells, and the random integration frequency was determined as previously described. The ratio of random integration frequency (the value of exon-trap vector is taken as 1) is shown. *: Not tested (an estimated value is presented, based on the assumption that random integration frequency is >10−3 in HeLa cells).
Before designing an exon-trap vector, the expression of the target gene must be confirmed in the cell line to be used. This may be determined by analyzing data from gene expression array analyses. If gene expression array data is unavailable, reverse transcription (RT)-PCR and/or Western blot should be performed to examine the gene expression levels.
Once the cellular expression of the target gene is confirmed, a gene-targeting strategy must be formulated, which includes determining the genomic region to be deleted and deciding the position and length of homology arms. The data relevant to this process may be obtained from the University of California Santa Cruz’s (UCSC) Genome Browser, and tips for designing exon-trap vectors are summarized in Fig. 3.
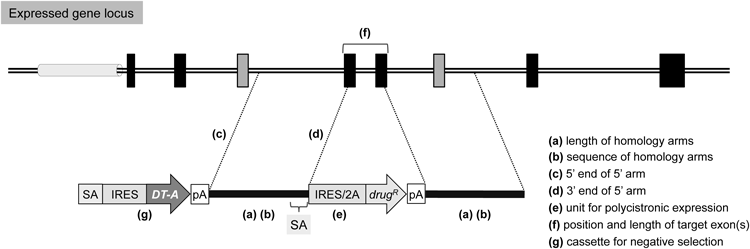
Exon-trap vectors are useful for genetically modifying expressed genes. The 5′- and 3′-homology arms should be designed as long as possible to achieve high-efficiency gene targeting (a), although longer arms can enhance random integration, presumably in a repetitive sequence-dependent fashion, and thus needs caution. For the same reason, repetitive sequences should be excluded from the arms when possible (b). The 5′ arm should not be set on or near the promoter region (c), since the presence of a promoter in the 5′ arm would hamper the strategy of exon-trapping. Additionally, disruption of a promoter region may affect nearby gene expression. In exon-trap vectors, a splice acceptor (SA) site should be attached upstream of a drug-resistance gene cassette. This SA site can be easily derived from an intron-exon boundary of the target gene, when the 3′ end of the 5′ arm is designed to locate on the exonic sequence (d), as shown in Fig. 4. The choice between IRES and 2A peptide sequences may depend on the expression level of the target gene (e), although the construction of 2A peptide-based vectors requires more effort than that of IRES-based vectors. Ideally, the genomic region to be deleted should be as large as possible to ensure the gene disruption; however, deleting the entire region is not recommended, for example, by the reason of the possible presence of non-coding RNA within an intron of the target gene. For additional reasons, the target region should be as short as possible (2 kb or less); additionally, the total number of nucleotides of the target exon(s) should be “3N±1,” not a multiple of three (f). In other words, the two exons (shown in gray) flanking the target exon(s) should not be in frame, for the purpose of avoiding the possibility of unexpected expression of truncated proteins. Finally, a negative selection cassette composed of a promoterless DT-A gene with an SA site and an IRES sequence is useful for reducing random integration frequency, when attached upstream of the 5′ arm of the vector (g). Symbols are as in Fig. 2. See the text for details.
The first and most critical step involves selection of the target region. Removal of the exon carrying the start codon may not be suitable if this exon is located in the vicinity of the promoter region, since modifying this region may affect the expression of the neighboring gene (or nearby genes).35) Gene disruption could be achieved by deleting the entire region (i.e., all exons); however, this strategy may lead to a reduction in targeting efficiency (our unpublished observations). Furthermore, deleting the entire region of the target gene may result in the simultaneous removal of additional genes (e.g., unknown non-coding RNA genes) located in the same region. Consequently, an exon(s) that are not located in the vicinity of the promoter region is chosen as the target region. Additionally, the target region should be as short as possible (2 kb or less). An additional consideration when determining the target region is the possibility of unexpected expression of truncated proteins (which may exert dominant negative effects). If the total number of nucleotides of the target exons is a multiple of three, the reading frame will not be altered even if these exons are skipped during splicing, thereby potentially leading to the expression of truncated proteins lacking the amino acid residues corresponding to the target exons. Therefore, an important criterion for determining the target region is that the two exons flanking the target exon should not be in frame.
As described in Section 3.1, longer homology arms are associated with higher targeting efficiency, even though the frequency of random integration increases in proportion to the length of repetitive DNA sequences present in the arms.20) We therefore suggest that homology arms should ideally contain as few repetitive sequences as possible. In Nalm-6 cells, gene targeting is feasible even with the use of vectors with 4-5-kb homology arms.36) However, these vectors are not suitable for the generation of homologous recombinants in human iPS cells or other human cell lines (our unpublished observations and ref. 37). Therefore, we propose that for gene targeting in human cells, the homology arms of targeting vectors should be ca. 9 kb or more in length, as reported by other groups.37,38)
Once the targeting strategy has been decided, the next step is vector construction. Following PCR amplification of each homology arm, targeting vectors may be constructed by standard molecular biology techniques utilizing restriction enzymes and DNA ligase. The use of standard techniques, however, is time- and labor-intensive as these involve cloning and mapping of DNA fragments. The MultiSite Gateway System (Life Technologies) enables the construction of exon-trap vectors in a much shorter time frame of only one week, as illustrated in Fig. 4. The major advantage of this system is that it does not require restriction mapping or ligation steps. Further, this method greatly facilitates construction of 2A peptide-containing exon-trap vectors, which are difficult to construct by conventional methods as stated above.34) The details of the vector construction system, transfection methods, and screening of homologous recombinants, have been described elsewhere.17,34) In addition to the MultiSite Gateway System, recently developed cloning methods such as In-Fusion Cloning (Clontech) or NEBuilder HiFi DNA Assembly (New England Biolabs) are also useful for the construction of exon-trap vectors. These methods enable the assembly of multiple DNA fragments by attaching a 15-nt homologous sequence to the 5′ end of the primers used to amplify the arms, thereby facilitating the vector construction in approximately the same time frame as with the MultiSite Gateway System.
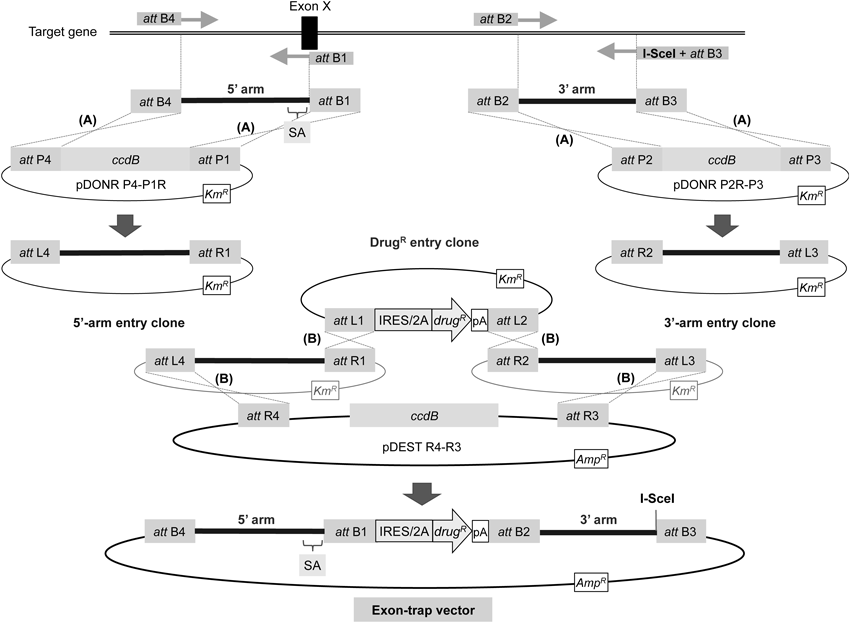
The 5′- and 3′-homology arms are PCR amplified with attB-containing primers, followed by generation of 5′- and 3′-arm entry clones via BP recombination reaction (denoted by A). The four attB sequences (attB4, attB1, attB2, and attB3) differ from one another, enabling efficient site-specific recombination. The reverse primer for 5′-arm amplification is set on an exon to be trapped (Exon X), naturally incorporating a splice acceptor site (SA) into the 5′ arm directly upstream of the drug-resistance gene cassette. Additionally, the presence of a unique restriction site (I-SceI in the figure) in the reverse primer for 3′-arm amplification enables linearization of the resultant targeting vector. The targeting vector is constructed by performing LR recombination reaction between four plasmids (denoted by B): the 5′- and 3′-arm entry clones obtained from BP recombination, an entry clone (DrugR entry clone) carrying an IRES/2A-linked drug-resistance gene flanked with attL1 and attL2 sequences,30) and the pDEST R4-R3 vector (Life Technologies). Symbols are as in Fig. 2. KmR, kanamycin-resistance gene; AmpR, ampicillin-resistance gene. ccdB, the gene encoding a DNA gyrase inhibitor, which allows for counterselection of nonrecombinant plasmids in E. coli. See refs. 17 and 34 for details.
Recently developed methods using artificial nucleases, such as TALEN or CRISPR/Cas, have improved genome-engineering technologies.3,4) TALENs comprise a FokI nuclease domain fused to a customizable DNA-binding domain, which is composed of highly conserved repeats derived from transcription activator-like effectors (TALEs) of the plant pathogenic bacterial genus Xanthomonas.39) CRISPR-Cas is a microbial adaptive immune system that uses RNA-guided nucleases to cleave foreign genetic elements. The Cas9 nuclease from Streptococcus pyogenes, which is the most commonly used Cas nuclease, can be guided by an engineered guide RNA (gRNA) designed to anneal to a target genome sequence of interest.40) These artificial nucleases (TALEN and Cas9) introduce a DSB at the target gene locus, thereby greatly increasing the gene-targeting efficiency when combined with exon-trap vectors.3,40) Indeed, by using a combination of CRISPR/Cas9 and exon-trap vectors, we have successfully obtained targeted clones in human iPS cells with >50% targeting efficiency (unpublished data). However, that such nuclease-mediated gene targeting may cause mutations due to unexpected off-target effects.5–8) Therefore, the development of new strategies compatible with exon-trapping targeting vectors is important for further improving gene-targeting technology.
Despite its utility, exon trap-based gene targeting is associated with two fundamental challenges. Firstly, expression of the positive selection marker not only occurs in correctly targeted clones, but also in random integrants where the vector has been inserted into a transcriptionally active non-target gene. Secondly, the exon trap-based method cannot be applied to genes with low expression levels. It is anticipated that resolution of these issues will render exon-trap vectors even more effective.
5.1 Development of a Novel Negative Selection CassetteWe recently developed a new cassette for negative selection, which is composed of a promoterless DT-A gene with an SA site and an IRES sequence. When this cassette is attached upstream of the 5′ arm of an exon-trap vector, which we termed ExTraPANS (exon-trapping positive and negative selection), random integrants in which the vector is inserted into a transcriptionally active non-target locus should be selectively eliminated by virtue of stable DT-A expression in these cells (Fig. 2). To confirm this, we employed ExTraPANS vectors for HPRT gene targeting in human HT1080 cells and mouse ES cells. As expected, the random integration frequency of ExTraPANS vectors was ca. 20% that of exon-trap vectors, resulting in a 5-fold higher gene-targeting efficiency34) (Table 1). Similarly, with the use of ExTraPANS vectors, the frequency of random integration in human iPS cells was decreased more than 4-fold, and the targeting efficiency was elevated to as high as 10% (Table 1 and our unpublished results). It should be noted that no increase in cytotoxicity was observed with ExTraPANS vectors, unlike in vectors with a promoter-containing DT-A gene,34) suggesting that transient expression of the promoterless DT-A gene does not occur upon transfection. These data indicate that this novel strategy using a promoterless DT-A gene cassette, which can efficiently counterselect random integrants, should greatly facilitate exon-trapping gene-targeting technologies.
5.2 Selective Induction of Target-Gene ExpressionWhen using exon-trap vectors, the expression of the drug-resistance gene is dependent on promoter activity of the target gene. Consequently, these vectors are not suitable for targeting genes with low expression levels. However, the application of exon-trap vectors for targeting genes with low expression levels may be achieved by activating the expression of the target gene via co-expression of TAL-VP64, which is a fusion protein composed of TAL (an effector domain of TALEN) and VP64 (a transcriptional activator composed of four tandem copies of the Herpes Simplex Viral Protein 16 domain (DALDDFDLDML),41,42) as illustrated in Fig. 5. The TAL-VP64 fusion protein lacks nuclease activity, but retains targeted DNA-binding activity and is expected to possess the ability to transcriptionally activate target gene expression. Indeed, we successfully used a TAL-VP64 fusion protein designed to bind the promoter region of a low-expression gene, thereby increasing the cellular expression of this gene (unpublished data). Advances in the development of methods for controlling target gene expression, such as those utilizing a nuclease-dead Cas9 protein (which lacks nuclease activity but retains DNA-binding capacity),43–45) are expected to increase the utility of exon-trap vectors for modification of non-expressed genes in the near future. It is important to mention, however, that these strategies should be based on the premise that artificial transcriptional activators do not disturb off-target gene expression.
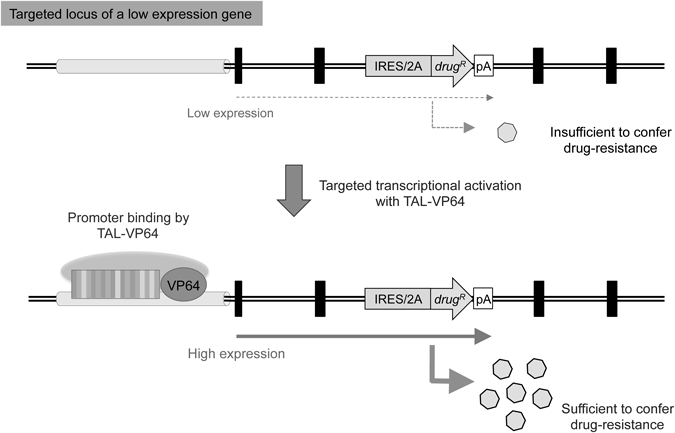
A promoterless drug-resistance gene is not applicable for the modification of genes with low expression levels, as the target gene must be expressed at a sufficient level to confer drug resistance. However, when an artificial transcriptional activator such as a TAL-VP64 fusion protein designed to bind the promoter region of a target gene is expressed in the cell, the genetic modification of low-expression genes becomes feasible (see the text for details). In such a strategy, however, unwanted side effects (i.e., perturbation of off-target gene expression or a possible genomic integration of the expression vector itself) should be carefully monitored. Symbols are as in Fig. 2.
In this review, we focused on the effect of vector structure on integration frequencies and on the effectiveness of exon-trap vectors. We also described a novel strategy, ExTraPANS, for negative selection, aimed at enhancing gene targeting by virtue of dramatically reduced random-integration frequencies. However, there is clearly still room for improvement. We encountered a considerable number of random integrants even when ExTraPANS vectors were used. Therefore, in addition to developing artificial nucleases with increased specificity, the development of targeting vectors capable of greatly suppressing the occurrence of random integrants is important for establishing safe and efficient gene-targeting techniques, particularly for therapeutic purposes.
We thank Aya Kurosawa, Yuta Abe, and Haruka Watabe for helpful discussions. This work was supported by Grants from Yokohama City University (Strategic Research Promotion S2501–S2601) and by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
The authors declare no conflict of interest.