2021 年 44 巻 11 号 p. 1617-1634
2021 年 44 巻 11 号 p. 1617-1634
The CYP3A subfamily, which includes isoforms CYP3A4, CYP3A5, and CYP3A7 in humans, plays important roles in the metabolism of various endogenous and exogenous substances. Gene and protein expression of CYP3A4, CYP3A5, and CYP3A7 show large inter-individual differences, which are caused by many endogenous and exogenous factors. Inter-individual differences can cause negative outcomes, such as adverse drug events and disease development. Therefore, it is important to understand the variations in CYP3A expression caused by endo- and exogenous factors, as well as the variation in the metabolism and kinetics of endo- and exogenous substrates. In this review, we summarize the factors regulating CYP3A expression, such as bile acids, hormones, microRNA, inflammatory cytokines, drugs, environmental chemicals, and dietary factors. In addition, variations in CYP3A expression under pathological conditions, such as coronavirus disease 2019 and liver diseases, are described as examples of the physiological effects of endogenous factors. We also summarize endogenous and exogenous substrates metabolized by CYP3A isoforms, such as cholesterol, bile acids, hormones, arachidonic acid, vitamin D, and drugs. The relationship between the changes in the kinetics of these substrates and the toxicological effects in our bodies are discussed. The usefulness of these substrates and metabolites as endogenous biomarkers for CYP3A activity is also discussed. Notably, we focused on discrimination between CYP3A4, CYP3A5, and CYP3A7 to understand inter-individual differences in CYP3A expression and function.
CYP3A plays important roles in the metabolism of various endogenous and exogenous substances. The CYP3A subfamily comprises isoforms CYP3A4, CYP3A5, CYP3A7, and CYP3A43 in humans (Table 1). CYP3A4 is the most important drug-metabolizing enzyme that metabolizes various substances. CYP3A5 is considered an enzyme comparable to CYP3A4 because of its high protein identity (84.26%, Table 1) and overlapping substrates with CYP3A4. Although these enzymes are often grouped together as CYP3A4/5 in many studies, differences between CYP3A4 and CYP3A5 have been disputed. The contribution of CYP3A4 and CYP3A5 to metabolism varies markedly depending on the substrate. For example, the intrinsic clearance of testosterone 6β-hydroxylation for CYP3A4 is higher than that for CYP3A5, while the intrinsic clearance of midazolam 1′-hydroxylation for CYP3A5 is 1 to 3-fold higher than that of CYP3A4.1,2) In addition, CYP3A4- or CYP3A5-specific substrates and inhibitors have been reported.3–6) Thus, the functions of CYP3A4 and CYP3A5 should be discriminated. CYP3A7 also has a high protein identity to CYP3A4 (88.47%) and CYP3A5 (81.87%, Table 1). CYP3A7 widely overlaps substrates with CYP3A4 and CYP3A5, although a few CYP3A7-specific substrates have been reported.7,8) CYP3A7 is the predominant isoform in the fetus and contributes to the metabolism of hormones and drugs in the fetus during pregnancy.7,9,10) CYP3A7 is found in some adult human liver, although CYP3A7 expression is decreased after birth.11) Therefore, it is necessary to discriminate between CYP3A4, CYP3A5, and CYP3A7 and clarify their expression and function. CYP3A43 is a minor isoform which function and clinical importance are not well understood and are not mentioned in this review.
| Symbol | Major regulatory factors (Nuclear receptors) | Protein identity (%)a) | Major expression tissues | Characteristics | References | |||
|---|---|---|---|---|---|---|---|---|
| RefSeq accession number | CYP3A4 | CYP3A5 | CYP3A7 | |||||
| Human | ||||||||
| CYP3A4 | PXR, CAR, VDR, FXR, PPARα, GR, ERα, LXR, HNF4α | NP_059488 | 100 | 84.26 | 88.47 | Liver, Small intestine, Kidney | Most abundant CYP3A isoform in human liver | 12,27–33,51,52,55) |
| CYP3A5 | PXR, CAR, GR, HNF4α | NP_000768 | — | 100 | 81.87 | Liver, Small intestine, Kidney, Lungs, Prostate | 29,34–36) | |
| CYP3A7 | PXR, CAR, GR | NP_000756 | — | — | 100 | Liver | Fetal dominant form | 37,38) |
| CYP3A43 | —b) | NP_073731 | 75.75 | 75.55 | 71.37 | Liver | 39) | |
| Mouse | ||||||||
| Cyp3a11 | PXR, CAR, HNF4α | NP_031844 | 72.62 | 72.96 | 70.04 | Liver, Small intestine | Most abundant CYP3A isoform in mouse liver Male dominant form | 40,41) |
| Cyp3a13 | CAR | NP_031845 | 75.65 | 74.35 | 72.46 | Liver, Small intestine | 42) | |
| Cyp3a16 | HNF4α | NP_031846 | 70.44 | 69.98 | 68.06 | Liver, Small intestine | Fetal dominant form | 43) |
| Cyp3a25 | CAR, HNF4α | NP_062766 | 71.17 | 71.12 | 67.40 | Liver, Small intestine | 41,42) | |
| Cyp3a41 | HNF4α | NP_001098629 | 71.23 | 71.77 | 68.65 | Liver | Female-specific form | 41,44) |
| Cyp3a44 | PXR, CAR, HNF4α | NP_796354 | 69.44 | 69.98 | 66.87 | Liver, Small intestine | Female-specific form | 40,41) |
| Rat | ||||||||
| Cyp3a1/23 | PXR, CAR, VDR, FXR | NP_037237 | 72.02 | 71.77 | 69.05 | Liver | Main CYP3A isoform in rat liver | 66) |
| Cyp3a2 | FXR | NP_695224 | 71.63 | 70.38 | 68.85 | Liver | Male-specific form | 66) |
| Cyp3a9 | PXR, GR | NP_671739 | 76.54 | 75.10 | 72.96 | Liver, Small intestine | Female dominant form | 45,66) |
| Cyp3a18 | —b) | NP_665725 | 68.41 | 68.61 | 65.19 | Liver, Small intestine | Male-specific form | 45) |
| Cyp3a62 | —b) | NP_001019403 | 71.63 | 73.44 | 69.82 | Small intestine | Female dominant form | 46) |
a): Analyzed using BLAST in NCBI, b): no significant reports, PXR: pregnane X receptor, CAR: constitutive androstane receptor, VDR: vitamin D receptor, FXR: farnesoid X receptor, PPARα: peroxisome proliferator-activated receptor α, GR: glucocorticosteroid receptor, ERα: estrogen receptor α, LXR: liver X receptor, HNF4α: hepatocyte nuclear factor 4α.
The gene or protein expression of CYP3A4, CYP3A5, and CYP3A7 shows large inter-individual differences in humans. In the adult human liver, CYP3A4 is the most abundant, followed by CYP3A5 among the CYP3A isoforms.12) CYP3A4 shows the greatest inter-individual differences, several tens to hundred-fold, in mRNA and protein expression in the liver.11,12) CYP3A5 also shows several to hundred-fold inter-individual differences in protein expression in the liver.2,11) Some adult livers express CYP3A7, while others have little.11) In the fetal human liver, CYP3A7 mRNA expression is the most abundant, followed by CYP3A5 (approximately 100-fold lower than CYP3A7) and CYP3A4 (approximately 1000-fold lower than CYP3A7).13) CYP3A7 expression shows several hundred-fold inter-individual differences.13) CYP3A5 expression also varies by several to hundred-fold.13,14) CYP3A4 mRNA expression shows ten-fold variation and increases with post-conception age.13,15) The inter-individual differences in the expression of CYP3A4, CYP3A5, and CYP3A7 are relevant to the differences in CYP3A function, which causes adverse outcomes such as disease and adverse drug events. Inter-individual differences are caused by various factors, such as genetic polymorphisms, age, sex, diseases, drugs, and environmental chemicals. Genetic polymorphisms, a congenital factor, are major factors for inter-individual differences in CYP3A expression and function. Some of the most common alleles for each CYP3A isoforms are CYP3A4*22, CYP3A5*3, and CYP3A7*1C. These variants have been reported to affect the metabolism of drugs and endogenous hormones.16–18) Genetic polymorphisms are especially described for CYP3A5,19) and CYP3A5*3 results in the loss of protein expression. In patients with CYP3A5*3/*3, the blood concentration of drugs, such as tacrolimus and apixaban, was higher than that in patients with CYP3A5*1/*1 (wild type). The relationship between CYP3A5 polymorphisms and tacrolimus pharmacokinetics has been well summarized, and the association between increase in the blood concentration of tacrolimus and adverse effects, especially renal dysfunction, has been discussed.20,21) The effects of CYP3A5 polymorphisms on apixaban pharmacokinetics have been recently reported,22,23) and an increase in apixaban exposure is associated with bleeding events.24) However, it is necessary to know both the congenital and acquired factors since congenital factors alone cannot explain the inter-individual differences in CYP3A expression and function. Changes in the gene and protein expression of CYP3A by acquired factors are directly linked to changes in CYP3A function. CYP3A4 mRNA expression is highly correlated with protein expression and enzyme activities.11,25,26) CYP3A5 and CYP3A7 show moderate correlations between mRNA and protein expressions.11) Hence, it is important to clarify the variations in gene or protein expression levels of CYP3A by various acquired factors to understand the inter-individual differences in CYP3A enzymatic activity.
In this review, we describe variations in gene and protein expression of CYP3A caused by endogenous and exogenous factors, and variations in the metabolism and kinetics of related endogenous and exogenous substrates. The relationship between variations in CYP3A expression and negative outcomes in our body (e.g., adverse drug events and diseases) and perspectives are discussed (Fig. 1).
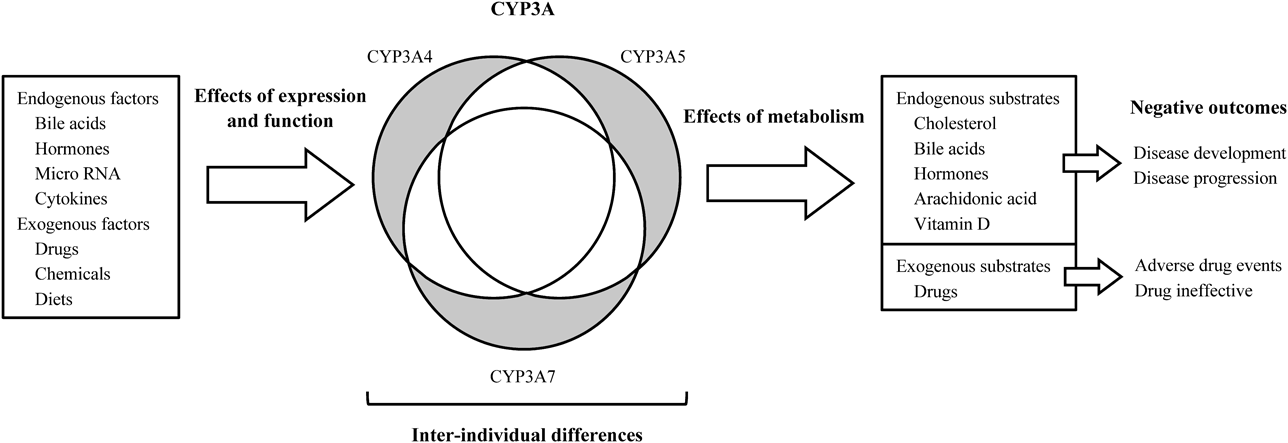
Gray areas mean isoform-specific characteristics.
CYP3A4, CYP3A5, and CYP3A7 expression are regulated and varied by various endogenous factors. These gene expressions are regulated and activated by the nuclear receptors listed in Table 1. Substances that activate or deactivate these nuclear receptors can alter CYP3A expression. Species differences have been demonstrated in studies of drug metabolism. Table 1 shows each CYP3A isoform in humans, mice, and rats. The expression levels and profiles, regulation, and function of CYP3A isoforms exhibit species-specific differences. In the research of drug metabolism, humanized animal models, including chimeric mice transplanted with human hepatocytes47) and human CYP3A4-transgenic mice,48) have been used, and the models also provide essential information about species differences.
In this section, major endogenous factors related to regulation and variation in CYP3A expression are summarized. Additionally, we describe details of the variation in CYP3A expression and related outcomes during pathological conditions in which the amount of endogenous factors increases or decreases.
2.1. Bile AcidsBile acids are activators of several nuclear receptors. Farnesoid X receptor (FXR) activation by major bile acids, such as cholic acid (CA), chenodeoxycholic acid (CDCA), lithocholic acid (LCA), deoxycholic acid (DCA), and ursodeoxycholic acid (UDCA), is summarized.49) CDCA is the most potent ligand of FXR, followed by DCA and LCA.50,51) FXR activation is considered to be involved in the induction of CYP3A4 expression.52) However, it has been suggested that FXR activation represses CYP3A4 expression by inducing FXR target genes which repress the transcriptional activation, a small heterodimer partner in the liver53) and fibroblast growth factor in the small intestine.54) In addition, bile acids activate pregnane X receptor (PXR). PXR activation induces the expression of CYP3A4, CYP3A5, and CYP3A7. LCA is the most potent ligand of PXR among natural bile acids, followed by DCA and CA.51) The vitamin D receptor (VDR) is also activated by LCA.51,55) VDR activation induces CYP3A4 predominantly in the small intestine.56) Bile acids are synthesized from cholesterol in the liver, which are called primary bile acids (including CA and CDCA), and secondary bile acids (including LCA, DCA, and UDCA) are produced by intestinal bacteria during enterohepatic circulation. Hence, it is suggested that the activation of these nuclear receptors by bile acids alters CYP3A gene expression in the liver and small intestine. LCA is known to induce CYP3A4 expression in the liver and intestine.57–59) Germ-free mice and antibiotic-treated mice showed low Cyp3a11 (generally comparable to CYP3A4) gene and protein expression in the liver, which is considered due to reduced production of LCA by intestinal bacteria.60,61) CA-fed mice showed induction of Cyp3a11 mRNA and protein expression in the liver.62–64) Schuetz et al. reported that this induction occurred in an FXR-dependent manner,62) whereas Zollner et al. reported that this occurred in an FXR-independent manner.63) However, CA has been reported to decrease CYP3A4 mRNA expression in primary human hepatocytes after 48 h of incubation.53) Although CDCA decreased the gene expression of CYP3A4 in human primary hepatocytes after 48 h of incubation,65) CDCA induced mRNA expression of CYP3A4 in human liver slices after 24 h of incubation.66) UDCA-fed mice induced Cyp3a11 mRNA expression in the liver in an FXR-independent manner.63) However, no changes in CYP3A4 mRNA and protein expression were observed in the human duodenum after daily UDCA intake for 3 weeks in healthy volunteers and patients with primary biliary cirrhosis.67) Therefore, the regulation of CYP3A expression by bile acids and the underlying mechanisms remain controversial. Bile acids, which can increase and decrease CYP3A4 expression, are likely involved in the homeostasis of CYP3A4 expression depending on the condition.
2.2. HormonesSeveral hormones, such as thyroid hormone (TH), parathyroid hormone (PTH), and growth hormone (GH), have the potential to modulate CYP3A expression. Triiodothyronine (T3), the active form of TH, has been reported to decrease the mRNA expression, protein expression, and activity of CYP3A4 in human primary hepatocytes.68) T3 administration for 2 weeks in humans decreased CYP3A enzymatic activity, as measured by the area under the plasma concentration–time curve (AUC) ratio for 1′-hydroxymidazolam/midazolam and the urinary ratio of 6β-hydroxycortisol/cortisol.69) Thyroid hormone receptors α and β, which are activated by TH, have been suggested to repress PXR-mediated gene expression of CYP3A4.70) PTH also reduced the mRNA and protein expression of Cyp3a2 in rats and CYP3A4 in human hepatocytes and Caco-2 colon carcinoma cells.71) GH has been suggested to contribute to the induction of CYP3A4 expression in human hepatocytes,68,72) and GH administration increased CYP3A4 activity, as measured by erythromycin breath tests in humans.73) In addition, GH has been shown to alter CYP3A expression in rats,74) and increase the expression of female-specific isoforms, Cyp3a41 and Cyp3a44, in mice.75) Continuous GH treatment stimulated liver CYP3A4 expression and activity in CYP3A4 transgenic male mice.76) GH might be the mechanism of sex differences in CYP3A expression because its secretory patterns are different between males and females.73,76,77) In fact, CYP3A4 expression in the female human liver is 1 to 2-fold higher than that in the male liver.12,78) The expression of CYP3A5 has also been reported to be sex-dependent in human hepatocytes.79) In addition to sex, hormone levels vary in a complex manner depending on the growth stage and life events, such as pregnancy and illness. Therefore, multiple hormones may combine to provide inter- and intra-individual differences in CYP3A basal expression and function.
A synthetic glucocorticoid, dexamethasone, is known to induce the expression and activity of CYP3A4, CYP3A5, and CYP3A7.80,81) In the fetal liver of CYP3A4/7-human PXR transgenic mice, it was suggested that endogenous glucocorticoids developmentally regulate CYP3A7 expression.82) Cortisol induced CYP3A4 and CYP3A5 expression and activity,72,83,84) and the induction is increased when cortisol is combined with other hormones, such as progesterone, estrogen, and GH, in pregnancy-mimic in vitro studies.72,83,85) Pregnancy is known to increase CYP3A activity in humans.86,87) Plasma concentrations of cortisol, estrogen, progesterone, and GH are markedly higher in pregnant women than in non-pregnant women.83) Female hormones, estrogen and progesterone, have also been reported to contribute to CYP3A induction during pregnancy. The mRNA expression levels of CYP3A4, but not CYP3A5, were increased by pregnancy-related concentration of estradiol, whereas both CYP3A4 and CYP3A5 mRNA expression levels were increased by progesterone in primary human hepatocytes.88) However, estrogen replacement therapy for menopause did not affect intestinal and hepatic CYP3A activity, as measured by the clearance of midazolam.89) These results suggest that the female hormonal effects on CYP3A expression may be pregnancy-specific.
2.3. Micro RNA (miRNA)MiRNAs are small non-coding RNAs with 21–25 nucleotides that act as posttranscriptional regulators. MiRNA 27b (miR-27b) has been reported to directly bind to the 3′-untranslated region (3′-UTR) of CYP3A4 and decrease CYP3A4 protein expression in vitro.90) miR-27b levels in blood negatively correlate with CYP3A4 activity in humans, suggesting that miR-27b is a useful biomarker.91,92) miR-543 directly binds to the 3′-UTR of CYP3A5 and decreases CYP3A5 protein expression.93) Indirect regulation of CYP3A4 by miRNAs, including miR-27b, has also been reported. miR-27b binds to VDR, resulting in a decrease in CYP3A4 protein expression.90) miR-148a affects the inducible and constitutive levels of CYP3A4 by decreasing the expression of PXR.94) In addition, it has been suggested that downregulation of CYP3A4 in the inflammatory state is associated with increase in several miRNAs due to inflammation.95) However, the effects of miRNAs on the expression of CYP3A5 and CYP3A7 are limited. Research on miRNAs has progressed dramatically in the last decade. Therefore, further studies are required to clarify the regulation of CYP3A4, CYP3A5, and CYP3A7 expression by miRNA.
2.4. Inflammatory CytokinesThe regulation of CYP by inflammation has been studied extensively, and the key factor of this mechanism is cytokines. The major cytokines, interleukin-1β (IL-1β), interleukin-6 (IL-6), and tumor necrosis factor α (TNF-α), downregulate CYP3A4 and CYP3A5 genes in human hepatocytes,96,97) Cyp3a11 and Cyp3a13 in mouse hepatocytes,98) and Cyp3a1 in rat hepatocytes.99) The regulation of CYP3A7 by inflammatory cytokines is not well understood. The downregulation of CYP3A is mediated by nuclear factor-kappa B (NF-κB) activation.100) In addition, TNF-α suppresses PXR-mediated transcriptional activation caused by rifampicin in in vitro studies,100,101) although in vivo reports on the effects of inflammation on CYP induction and transcriptional activation of nuclear receptors are limited. The effects of inflammation on changes in pharmacokinetics and related adverse effects are widely suggested in patients with inflammation, such as liver transplantation,102) Crohn’s disease,103) and coronavirus disease 2019 (COVID-19).104–107) Details of the variation in CYP3A expression and related outcomes in patients with COVID-19 and under conditions of major liver disease and related surgery are described below.
2.5. Variation in CYP3A Expression and Related Outcome during Pathological Conditions2.5.1. COVID-19COVID-19, caused by infection with novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a recent pandemic of global concern. The inflammatory response is associated with COVID-19. The levels of cytokines and C-reactive protein (CRP) are increased, and inflammatory biomarkers are related to disease severity.108,109) The inflammatory state in patients with COVID-19 is regarded to affect the metabolism of therapeutic drugs. Although changes in CYP3A expression in patients with COVID-19 are not directly known, the effects of inflammation on the pharmacokinetics of CYP3A substrates have been evaluated, as shown in Table 2. Human immunodeficiency virus (HIV) protease inhibitors, such as darunavir, lopinavir, and ritonavir, are repurposed to treat COVID-19, and these therapeutic drugs are mainly metabolized by CYP3A4. The maximum and minimum blood concentrations of darunavir were higher (approximately 1.5- and 5-fold, respectively) in patients infected with SARS-CoV-2 than in patients infected with HIV after administering the same dosage of darunavir. In the population pharmacokinetic analysis of darunavir, IL-6 is suggested to be the most significant clinical covariate affecting the oral clearance of darunavir.104) The combination of lopinavir and ritonavir is used because ritonavir is a competitive inhibitor of lopinavir metabolism, resulting in an increased blood concentration of lopinavir. The trough blood concentration of lopinavir in patients infected with SARS-CoV-2 is 3 to 4-fold higher than that in HIV-infected patients after administering the same dosage of lopinavir and ritonavir.105–107) The blood concentration of lopinavir is positively correlated with CRP values.106,107) In contrast, no differences in the blood concentration of hydroxychloroquine were observed between patients infected with SARS-CoV-2 and control patients with systemic lupus erythematosus.107) Although the impact of the inflammatory state in patients infected with SARS-CoV-2 on pharmacokinetics may vary depending on the pharmacokinetic properties of drugs, the monitoring of pharmacokinetics is needed during pharmacotherapy.
| Drug | Major drug metabolizing enzymes | Patients and numbers | Inflammatory biomarkers | Administration | Pharmacokinetics | Comments | Adverse effects | References |
|---|---|---|---|---|---|---|---|---|
| Darunavir | CYP3A4 | SARS-CoV-2-infected patients n = 30 | IL-6: 31.0 pg/mL (10–114.75) | 800 mg/d with other drugs | Median Cmin: 4960.2 ng/mL (IQR 2015.0–7951.2) Median Cmax: 9476.2 ng/mL (IQR 7220.5–14835.4) | High Cmin and Cmax compared to HIV-infected patients (as below) | Abdominal pain/diarrhea (n = 3) QT prolongation (n = 2) Asthenia (n = 1) Tonic–clonic seizure (n = 1) Rash (n = 1) | 106) |
| HIV-infected patients n = 25 | IL-6: 2.0 pg/mL (2.0–2.75) | 800 mg/d with other drugs | Median Cmin: 1010.0 ng/mL (IQR 550.0–2112.0) Median Cmax: 6320.0 ng/mL (IQR 3888.3–7747.5) | Abdominal pain/diarrhea (n = 1) | ||||
| Lopinavir | CYP3A4 CYP3A5 | SARS-CoV-2-infected patients n = 11 | CRP: 48.9 mg/L (range 19.4 to 157.5) n = 10 | 400 mg with 100 mg ritonavir twice a day | Median Ctrough: 18000 ng/mL (range 11400 to 30800) | High Ctrough compared to HIV-infected patients (5365 ng/mL) | Diarrhoea (n = 6) Nausea and vomiting (n = 2) | 107) |
| Lopinavir Ritonavir | Lopinavir: CYP3A4 CYP3A5 Ritonavir: CYP2D6 CYP3A4 | SARS-CoV-2-infected patients n = 8 | CRP: 298 mg/L (183–353) | Lopinavir 400 mg Ritonavir 100 mg twice a day | Lopinavir Ctrough: 27908 ng/mL (IQR 15928–32627) Ritonavir Ctrough: 634 ng/mL (IQR 255–1269) | High Ctrough compared to upper limit of lopinavir Ctrough in HIV-infected patients (8000 ng/mL) Positive correlation between CRP and Ctrough of lopinavir | Cholestasis (n = 1) | 108) |
| SARS-CoV-2-infected patients n = 9 | CRP: 263 mg/L (157–327) | Lopinavir 400 mg Ritonavir 100 mg once a day | Lopinavir Ctrough: 22974 ng/mL (IQR 21394–32735) Ritonavir Ctrough: 186 ng/mL (IQR 15–474) | High Ctrough compared to upper limit of lopinavir Ctrough in HIV-infected patients (8000 ng/mL) | — | |||
| Lopinavir | CYP3A4 CYP3A5 | SARS-CoV-2-infected patients n = 92 | CRP: 65 mg/L (36–113) | Lopinavir 800 mg Ritonavir 200 mg twice on day 1 Lopinavir 400 mg Ritonavir 100 mg twice on and after day 2 | Median Ctrough: 26500 ng/mL (range 7700 to 42300) | High Ctrough compared to HIV-infected patients (7100 ng/mL) Low Ctrough in patients combined with tocilizumab (humanized monoclonal antibody against IL-6 receptor) compared to that in patients with no tocilizumab (18700 versus 28800 ng/mL) | — | 109) |
| CRP: <75 mg/L n = 52 | Median Ctrough: 20900 ng/mL | — | ||||||
| CRP: ≥75 mg/L n = 40 | Median Ctrough: 30700 ng/mL | High Ctrough compared to patients with CRP levels of <75 mg/L (as above) | — | |||||
| Hydroxy chloroquine | CYP2C8 CYP3A4 | SARS-CoV-2-infected patients n = 59 | CRP: 65 mg/L (IQR 36–113) | 800 mg once 400 mg after 6, 24, 48 h with lopinavir and ritonavir | Median Ctrough: 171 ng/mL (range 56 to 454) | Comparable to Ctrough in patients with systemic lupus erythematosus (103 to 130 ng/mL) | — | 109) |
| CRP: <75 mg/L n = 29 | Median Ctrough: 149 ng/mL | No correlation between CRP and Ctrough of hydroxychloroquine | — | |||||
| CRP: ≥75 mg/L n = 22 | Median Ctrough: 148 ng/mL | — |
IL-6: interleukin-6, CRP: C-reactive protein, Cmin: minimum blood concentration, Cmax: maximum blood concentration, Ctrough: trough blood concentration, IQR: interquartile range.
The liver is the most important organ for drug metabolism. Liver diseases, such as hepatitis, nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), and hepatocellular carcinoma (HCC), are known to affect CYP3A expression in humans and animals. NAFLD is a liver disease defined by the presence of steatosis, which is not caused by alcohol use, and its more progressive form NASH is associated with inflammation and fibrosis. The mRNA and protein expression of CYP3A4 in the liver is approximately 2-fold lower in human NAFLD and NASH donors compared to normal donors, and the enzymatic activity of CYP3A measured by midazolam or 4β-hydroxycholesterol concentration is also lower.110,111) In addition, the protein expression and activity of CYP3A4 are thought to decrease with the severity and progression of steatosis.111,112) However, the pharmacokinetic parameters of apixaban, which is a substrate for CYP3A4 and CYP3A5, did not change in patients with NAFLD.113) Clinical information on the pharmacokinetic properties of drugs in patients with NAFLD or NASH is limited. However, the changes in CYP3A expression were not consistent with the results obtained using animal models. Several reports have shown a decrease in the expression of CYP3A in high fat diet-induced NAFLD rodent models, whereas a few reports have shown an increase.110,114,115) The reason for this discrepancy is unclear. In patients with HCC, mRNA and protein expression of CYP3A4 in the liver is lower than that in the normal liver.116,117) The metabolism of sorafenib mediated by CYP3A4 and uridine diphosphate glucuronosyltransferase (UGT) 1A9 was markedly decreased in human tumor hepatic microsomes.118) The downregulation of the CYP3A4 gene and protein was correlated with a poor prognosis, suggesting that CYP3A4 levels are a useful biomarker in HCC.119) CYP3A5 expression is also downregulated in HCC tissues and negatively associated with metastasis.120) Thus, CYP3A4 and CYP3A5 can contribute to the development and progression of liver diseases as well as drug metabolism in patients with liver diseases.
Liver resection is performed to remove lesions in patients with liver cancer, and the healthy liver regenerates after surgery. CYP3A expression and related activities are regarded to be altered during liver regeneration. However, the variation in CYP3A expression during liver regeneration in humans is not clear. In mice and rats, mRNA or protein expression of CYP3A in the liver decreases immediately after liver resection and then gradually increases with regeneration.121–123) Furthermore, mRNA expression of Cyp3a13 (comparable to CYP3A5),124) increased in the small intestine of mice after liver resection and then gradually recovered with regeneration, suggesting compensatory induction instead of a decrease in CYP expression in the liver.123) Muraki et al. suggested that the increase in CYP3A5 activity in the small intestine after living-donor liver transplantation in humans, which is consistent with the results of Cyp3a13 expression in the small intestine of mice.125) It has been suggested that changes in drug metabolism in the small intestine and liver should be noted after liver resection or transplantation. In addition, these changes in CYP3A expression in liver disease and related surgery, as described above, are considered to be caused by complex factors, but the detailed mechanisms remain unelucidated. It is necessary to investigate the changes in CYP3A expression in response to changes in endogenous substances, including inflammatory cytokines, hormones, growth factors, and bile acids, which vary under these pathological conditions.
Various exogenous factors, such as drugs, environmental chemicals, and foods, are known to alter the expression of CYP3A. In this section, representative exogenous factors that can upregulate and downregulate CYP3A expression are described.
3.1. DrugsRifampicin is a typical PXR-mediated CYP3A4, CYP3A5, and CYP3A7 inducers in humans, but not in rodents. There are species differences in the ligand-dependent activation of nuclear receptors, including PXR and constitutive androstane receptor (CAR). However, the ability of rifampicin as a CYP3A inducer is clinically relevant and has been well reviewed for its impact on the kinetics of various drugs and endogenous substrates.126,127) Many marketed drugs, such as carbamazepine, dexamethasone, phenobarbital, and phenytoin, have been reported to induce CYP3A4 and CYP3A5 through the activation of nuclear receptors.128) To the best of our knowledge, specific inducers of CYP3A4, CYP3A5, and CYP3A7 have not been reported. The CYP3A induction potential has been investigated in drug development.129) In 2015, a total of 33 drugs were approved by the U.S. Food and Drug Administration (FDA), and 12 compounds and metabolites were found to have CYP3A induction potential in human hepatocytes.130) In 2017, a total of 34 drugs were approved, and 10 compounds and metabolites were found to have CYP3A induction potential.131) Several drugs have been shown to induce in vivo induction of CYP3A, associated with drug-drug interactions.130,131) In contrast, acetaminophen was reported to induce CYP3A1 protein expression with unchanged mRNA expression by inhibiting the degradation of CYP3A1 protein in rat primary hepatocytes.132,133) CYP3A isoforms and related nuclear receptors also undergo post-translational regulation, such as ubiquitination of CYP3A4134,135) and CYP3A5,135) phosphorylation of CYP3A4,136) and PXR,137) and miRNA (described in Section 2). Phosphorylation sites have also been reported in CYP3A7.138) Therefore, certain drugs can change CYP3A expression and function through post-translational regulation, as well as transcriptional activation, and affect the kinetics of various drugs and endogenous substrates. Therefore, it is necessary to understand the mechanisms of CYP3A induction.
The guidelines of Pharmaceuticals and Medical Devices Agency (PMDA) shows the necessity of in vitro enzyme induction and down-regulation studies during drug development to evaluate the possibility of drug–drug interactions (DDIs) (https://www.pmda.go.jp/files/000228122.pdf). Several drugs have been shown to downregulate CYP.139) For example, metformin, which is used to treat type-2 diabetes, dramatically suppressed PXR-mediated expression and induction of CYP3A4 in human hepatocytes and LS174T colon adenocarcinoma cells, as well as Cyp3a11 in the liver and intestine of mice.140) However, the information on down-regulation by other drugs in vivo and the significant clinical importance of CYP3A down-regulation by drugs to DDI remain unclear. Down-regulation by drugs is very likely to affect DDI because the clinical relevance of CYP3A downregulation by endogenous factors under pathological conditions has been reported, as described in Section 2. Further research on CYP3A down-regulation by drugs is expected using both in vitro and in vivo models.
3.2. Environmental ChemicalsChemicals contained in daily exposed products, such as foods, household goods, and personal care products, can affect CYP3A expression. Many pesticides, including pyrethroids, organophosphorus, and carbamates, have been reported as activators of PXR and CAR in humans and rodents, such as permethrin, deltamethrin and fenvalerate (pyrethroids), profenofos and chlorpyrifos (organophosphorus), benfuracarb, and carbaryl (carbamates).141–145) The related pesticides increased the mRNA expression levels and activity of human CYP3A4 in in vitro studies.143) There are species differences in PXR and CAR activation by several pesticides between humans and mice.142,143) In addition, several metabolites of pesticides showed different degrees of PXR activation compared to the parent compound.144,145) CYP3A5 mRNA expression was increased by permethrin in human hepatocytes, but not in deltamethrin.146) CYP3A7 mRNA expression was increased by imidacloprid,147) which belongs to the class of neonicotinoids and has been reported to activate PXR.148) However, the effects of other pesticides on the expression of CYP3A5 and CYP3A7 are unknown. It will be necessary to investigate the effects of pesticides on CYP3A5 and CYP3A7 expression to understand the changes in CYP3A function caused by pesticides.
Parabens (a homologous series of alcohol esters of 4-hydroxybenzoic acid), a kind of preservatives, have the potential to PXR, CAR, and peroxisome proliferator-activated receptor α (PPARα) activation in rats and human. Transcriptional activation differed by the side chain of parabens.149) Human and rat PXR and CAR were activated by parabens with C2–C5 linear and branched alkyl sidechains, whereas rat PPARα was activated by parabens with C7–C12 linear alkyl sidechains. In addition, rat PXR antagonistic activity and CAR inverse agonistic activity by pentylparaben and hexylparaben were observed. A metabolite of parabens, 4-hydroxybenzoic acid, did not activate PXR, CAR, and PPARα.149) However, to our knowledge, CYP3A induction and its related effects on the kinetics of endogenous and exogenous substrates have not been reported.
The PXR and CAR activities of other environmental chemicals have also been reported, such as benzotriazole UV stabilizers150) and plasticizers.151) The activation of PXR and CAR has been reported for a wide range of substances. It is considered that the ligand binding sites of PXR and CAR are flexible. Thus, chemicals that are routinely exposed may be associated with inter- and intra-individual variations in CYP3A expression and function, although further investigation is needed.
3.3. Dietary FactorsNatural compounds in the diet can alter CYP3A expression. Polyphenols, which include flavonoids, are found in plant foods such as fruits, vegetables, coffee, tea, herbs, chocolate, and wine. Polyphenols have been reported to have potential as CYP3A4 inducers in experiments using human hepatocytes or HepG2 hepatocarcinoma cells. For example, resveratrol, schisantherin A, schisandrin A, schisandrin B, schisandrol A,152) baicalein,153) apigenin, and luteolin154) activate PXR and increase CYP3A4 expression. Conversely, silybin and isosilybin showed PXR antagonistic activity, resulting in the inhibition of CYP3A4 induction in LS180 colon adenocarcinoma cells.155) The changes in CYP3A5 and CYP3A7 expression by polyphenols are not well known. On the other hand, inhibition of CYP3A enzymatic activity by polyphenols and contained foods has been extensively investigated.156–158) Further knowledge of the effects of dietary intake of these compounds on systemic metabolic potential in humans is needed.
In vivo studies using rodents, a type of diet, affect basal CYP gene expression. Feeding a purified diet (AIN-93G or AIN-93M), in which each nutrient is supplied by a predetermined and purified ingredient, reduced mRNA expression of Cyp3a11, Cyp3a13, and Cyp3a41 in the liver and small intestine of mice, compared to a standard diet containing natural cereal-based foods.123,159,160) In mice fed the purified diet, the plasma bile acid concentrations were lower than those in mice fed the standard diet, suggesting that bile acids partly cause these differences in basal CYP3A expression.159) Feeding a high-fat diet for a few weeks, significant changes in Cyp3a11, Cyp3a13, and Cyp3a41 mRNA expression were not observed in mice.160,161) However, feeding a high-fat diet for a few months causes steatosis or NAFLD, and CYP3A expression is altered in rodents.114,115) Thus, it seems that daily eating habits determine basal CYP3A expression and function, but understanding the effects of all human diets is difficult. Basal CYP3A expression and function should be predicted using biomarkers of CYP3A activity, as described below.
Changes in CYP3A expression can alter the kinetics of various endogenous substances, affecting homeostasis in the body. In this section, major endogenous substrates metabolized by CYP3A are described, and their usefulness as biomarkers for CYP3A activity and the effects of changes in CYP3A expression on kinetics, toxicity, and disease are discussed. Table 3 shows CYP3A-specific metabolites as expected biomarkers for CYP3A enzymatic activity, and their contributions and specificities of the three isoforms are summarized.
| Expected biomarkers of CYP3A activity | Parent compound | Contribution and specificitya) | References | |||
|---|---|---|---|---|---|---|
| CYP3A4 | CYP3A5 | CYP3A7 | Clinical reports | |||
| 4β-Hydroxycholesterol | Cholesterol | Yes | Yes (4 > 5) | — | 167) | 86, 162–164) |
| 25-Hydroxycholesterol | Cholesterol | Yes | Yes (4 > 5) | — | 167) | |
| 5β-Cholestane-3α,7α,12α,25-tetrol | 5β-Cholestane-3α,7α,12α-triol | Yes | Yes (4 > 5) | — | 170) | |
| 6α-Hydroxytaurochenodeoxycholic acid | Taurochenodeoxycholic acid | Yes | — | — | 171) | |
| 6α-Hydroxylithocholic acid | Lithocholic acid | Yes | — | — | 171) | |
| Dehydrolithocholic acid | Lithocholic acid | Yes | Yes (4 ≈ 5) | — | 172) | |
| 1β-Hydroxydeoxycholic acid | Deoxycholic acid | Yes | Yes | Yes (4 ≈ 7 > 5) | 8) | 175) |
| 3-Dehydrodeoxycholic acid | Deoxycholic acid | Yes | Yes | Yes (4 ≈ 7 > 5) | 8) | |
| 5β-Hydroxydeoxycholic acid | Deoxycholic acid | Yes | Yes | Yes (4 ≈ 7 > 5) | 8) | |
| 6β-Hydroxydeoxycholic acid | Deoxycholic acid | Yes | No | Yes (4 ≈ 7) | 8) | |
| 4β/6α-Hydroxydeoxycholic acid | Deoxycholic acid | Yes | No | Yes (7 > 4) | 8) | |
| 19-Hydroxydeoxycholic acid | Deoxycholic acid | No | No | Yes | 8,173) | |
| 6β-Hydroxycortisol | Cortisol | Yes | Yes | Yes (4 > 5 ≈ 7) | 10) | 177,178) |
| 6β-Hydroxycortisone | Cortisone | Yes | — | — | 180) | 181) |
| 6β-Hydroxytestosterone | Testosterone | Yes | Yes | Yes (4 > 5 > 7) | 1,2,7,10) | |
| 2β-Hydroxytestosterone | Testosterone | Yes | Yes | Yes (4 ≈ 5 > 7) | 7) | |
| 2α-Hydroxytestosterone | Testosterone | No | No | Yes | 7) | |
| 7β-Hydroxydehydroepiandrosterone | Dehydroepiandrosterone | Yes | Yes | Yes (4 > 5 ≈ 7) | 185,186) | 187) |
| 16α-Hydroxydehydroepiandrosterone | Dehydroepiandrosterone | Yes | Yes | Yes (7 > 4 > 5) | 185,186) | 187) |
| 7α-Hydroxydehydroepiandrosterone | Dehydroepiandrosterone | Yes | Yes (4 ≈ 5) | No | 186) | |
| 1,23R,25-Trihydroxycholecalciferol | 1,25-Dihydroxycholecalciferol | Yes | Yes (4 > 5) | — | 208) | |
| 4β, 25-Dihydroxycholecalciferol | 25-Hydroxycholecalciferol | Yes | — | — | 209) | 209) |
a) Yes: its isoform was reported to contribute to the metabolism, No: its isoform was reported not to contribute to the metabolism, —: no information, 4: CYP3A4, 5: CYP3A5, 7: CYP3A7, >: left isoform showed higher contribution (intrinsic clearance or equivalent) to the metabolism than right isoform, ≈: left and right isoforms showed similar contributions.
Cholesterol is metabolized to 4β-hydroxycholesterol by CYP3A4 and CYP3A5, and 4β-hydroxycholesterol is expected to be a useful biomarker for CYP3A activity. The plasma or urinary 4β-hydroxycholesterol/cholesterol ratio has been used to predict CYP3A activity in clinical studies,86,162–164) although some limitations need to be considered, for example, involvement of intestinal CYP3A4 in 4β-hydroxycholesterol formation.165,166) Cholesterol is also metabolized to 25-hydroxycholesterol by CYP3A4 and CYP3A5, and the concentration of 25-hydroxycholesterol in the liver and plasma was correlated with CYP3A4 protein levels in the liver of CYP3A-humanized mice.167) CYP3A4 showed a minor contribution to the 22-, 24-, 26-, and 27-hydroxylation of cholesterol.168) Hashimoto et al. suggested that Cyp3a is important for the homeostasis of cholesterol and bile acids in Cyp3a knockout mice.169) Cholesterol is converted to bile acids mediated by various enzymes, including CYP7A1, CYP27A1, CYP8B1, CYP3A4, and CYP3A5 in humans. The 25-hydroxylation of 5β-cholestane-3α,7α,12α-triol, which is an intermediate form of cholic acid formation, is catalyzed by CYP3A4 and CYP3A5.170) In bile acid homeostasis, CYP3A4 catalyzes 6α-hydroxylation of both taurochenodeoxycholic acid and LCA.171) LCA is metabolized to the 3-oxidation form by CYP3A4 and CYP3A5.172) In addition, DCA is metabolized to 1β, 3β, 4β, 5β, 6α, 6β, and 19-oxidation forms by CYP3A4 and CYP3A7.173,174) The urinary metabolic ratio of 1β-hydroxyDCA to DCA has been suggested as a potential CYP3A endogenous biomarker.175) 19-Hydroxylated DCA, which is specifically generated by CYP3A7, is suggested as an in vitro marker for CYP3A7 activity although the usefulness of this marker in vivo is unclear.8) In contrast, the deletion of Cyp3a showed no major differences in bile acid composition in Cyp3a knockout mice.176) Although CYP3A contributes to many metabolic pathways of bile acids, the importance of CYP3A in bile acid homeostasis needs to be clarified in detail.
4.2. HormonesCYP3A plays an important role in hormone homeostasis CYP3A4, CYP3A5, and CYP3A7 metabolize cortisol to 6β-hydroxycortisol.10) In clinical studies, the urinary metabolic ratio of 6β-hydroxycortisol to cortisol has been widely characterized as a CYP3A endogenous biomarker,177,178) although its use is susceptible to circadian variation.179) Cortisone is also metabolized to 6β-hydroxycortisone by CYP3A.180) Although little is known about the isoform specificity of 6β-hydroxylation, the urinary metabolic ratio of 6β-hydroxycortisone to cortisone has also been suggested as a biomarker for CYP3A activity.181)
The metabolism of sex steroids mediated by CYP3A has been well discussed. CYP3A4, CYP3A5, and CYP3A7 catalyze 6β-hydroxylation of testosterone, which are widely used as in vitro markers of CYP3A activity.182) CYP3A4 also mediates the 2β-, 15-, and 16β-hydroxylation of testosterone.183) CYP3A5 and CYP3A7 metabolize testosterone to 6β- and 2β-hydroxytestosterone, whereas 2α-hydroxytestosterone is formed by CYP3A7, but not by CYP3A4 and CYP3A5.7) However, the ratio of 2α-hydroxytestosterone to 6β-hydroxytestosterone is unlikely to be an effective endogenous biomarker for CYP3A7 activity because the ratio varies with testosterone concentration.7) In Cyp3a knockout mice, plasma testosterone levels were dramatically increased because of a decrease in the metabolism of testosterone, and androgen response was stimulated in the prostate.184) CYP3A4, CYP3A5, and CYP3A7 catalyze 7β- and 16α-hydroxylation of dehydroepiandrosterone (DHEA). The major metabolite of CYP3A4 is 7β-hydroxyDHEA, whereas the major metabolite of CYP3A7 is 16α-hydroxyDHEA.185,186) The profiles of the 16α- and 7β-hydroxylation forms can be used to differentiate CYP3A4 and CYP3A7 activity, and the contribution of CYP3A5 to these products is negligible.185) The 7α-Hydroxylation form of DHEA is produced by CYP3A4 and CYP3A5, but not by CYP3A7.186) In addition, a statistical prediction model was constructed by combining the ratios of 7β-hydroxyDHEA/DHEA, 16α-hydroxyDHEA/DHEA, and 6β-hydroxycortisol/cortisol.187) Estradiol is metabolized by CYP3A4 to form 2-, 4-, and 16β-hydroxylation. In human CYP3A4-transgenic mice, serum estradiol levels were significantly lower than those in wild-type mice during pregnancy and lactation because of an increase in the metabolism of estradiol.188) CYP3A5 and CYP3A7 also catalyze the 2- and 4-hydroxylation of estradiol, although their contributions are lower than those of CYP3A4.1) However, hydroxylation of estradiol is mainly mediated by another CYP subfamily, CYP1A.189,190) Moreover, CYP3A4, CYP3A5, and CYP3A7 contribute to catalyze the 2-, 4-, and 16α-hydroxylation of estrone.189–192) CYP3A4 catalyzes the 2β-, 6β-, 16α-, and 21-hydroxylation of progesterone.183) The 6β-hydroxylation of progesterone is also catalyzed by CYP3A5 and CYP3A7.10) Thus, CYP3A is an important enzyme that catalyzes the metabolism of sex steroids, suggesting an association with the development of hormone diseases, such as breast cancer and prostate cancer. In clinical studies, the expression and polymorphisms of CYP3A4, CYP3A5, and CYP3A7 have been reported to be associated with susceptibility to breast cancer18,193–195) and prostate cancer.196–198)
4.3. Arachidonic Acid CascadeArachidonic acid, which is freed from phospholipids of the cell membrane by phospholipase A2, is a precursor to various eicosanoids biosynthesized in three main pathways. The first pathway is the cyclooxygenase pathway, which biosynthesizes prostaglandins and thromboxane. The second pathway is the lipoxygenase pathway, which biosynthesizes leukotrienes and lipoxane. The third pathway is the CYP-mediated pathway, which biosynthesizes epoxyeicosatrienoic acid (EET) and hydroxyeicosatetraenoic acid (HETE). In the human CYP-mediated pathway, the major CYP subfamily that forms EET is CYP2C8, CYP2C9, CYP1A2, and CYP3A4 in the liver199,200) and CYP2J2 in the heart,201) and CYP4F2 is a major CYP subfamily that forms HETE in the liver and kidneys.200,202) CYP3A4 contributes to the metabolism of arachidonic acid to (±)-8,9-, (±)-11,12-, and (±)-14,15-EET in the liver.200) EET has various physiological functions, such as lowering blood pressure, anti-inflammatory action, and promoting cell proliferation203,204); therefore, variation in CYP3A4 expression can change EET levels and function. CYP3A4 knockdown in breast cancer cell lines significantly reduced EET content and blocked cancer cell proliferation.205) CYP3A4 overexpression promoted cell growth in Hep3B hepatocarcinoma cells, and cell growth was attenuated by an EET antagonist.206) Thus, these studies suggest that CYP3A4 contributes to tumor development and progression. In clinical studies, the expression and polymorphisms of CYP3A4 and CYP3A5 have been reported to be associated with susceptibility to lung cancer,207) breast cancer,193–195) and prostate cancer.196–198) However, little is known about the involvement of CYP3A5 and CYP3A7 in EET biosynthesis.
4.4. Vitamin DThe bioactive form of vitamin D, 1,25-dihydroxycholecalciferol (1,25(OH)2D3), and its precursor, 25-hydroxycholecalciferol (25(OH)D3), is metabolized by CYP3A4 in the liver and small intestine,208,209) although CYP24A1 also catalyzes the 23- and 24-hydroxylation of 1,25(OH)2D3 and 25(OH)D3, especially in the kidneys.210,211) CYP3A4 and CYP3A5 catalyze 23- and 24-hydroxylation of 1,25(OH)2D3 in the liver and small intestine.208) 1,23R,25(OH)2D3 and 4β,25(OH)2D3 are the CYP3A4-mediated major metabolites of 1,25(OH)2D3 and 25(OH)D3, respectively.209) However, 1,25(OH)2D3 has been reported to induce the expression and activity of CYP3A4 through VDR activation, especially in the small intestine.56) Vitamin D contributes to the transcriptional regulation of CYP3A4, which metabolizes vitamin D, suggesting that feedback control may occur to maintain vitamin D homeostasis.
Endogenous biomarkers for CYP3A activity are expected because these biomarkers can be measured with less invasiveness. Only urine or a single blood sample following biochemical tests is needed without the administration of xenobiotics. As shown in Table 3, these metabolite levels or the ratio of the metabolite to parent compound levels in blood or urine can predict inter-individual differences in CYP3A activity and isoform-specific activity. In addition, the combination of some markers leads to more accurate predictions of the activity under consideration of the contribution of each isoform, CYP3A4, CYP3A5, and CYP3A7. Accurate predictions of CYP3A activity are useful for providing effective pharmacological therapy and evaluating the development and progression of diseases, such as NAFLD and cancer.
Changes in CYP3A expression also alter the pharmacokinetics of various drugs, resulting in drug ineffectiveness or adverse effects during pharmacotherapy. More than 30% of marketed drugs are metabolized by CYP3A4 and CYP3A5. For example, 1′-hydroxylation of midazolam is specifically mediated by CYP3A and is widely used as a probe drug for CYP3A activity in vitro and in vivo. CYP3A7 has the potential to metabolize various drugs that overlap substrates with CYP3A4 and CYP3A5.1) Metabolism of drugs such as imipramine,212) glyburide,9) and oxycodone213) by CYP3A7 has been reported. CYP3A4 and CYP3A5 metabolize the largest number of new drugs recently approved by the FDA.130,131)
DDIs between CYP3A substrates and inducers are widely known. For example, a typical CYP3A inducer, rifampicin, decreases the blood concentration of various CYP3A substrates such as midazolam, triazolam, and simvastatin.126) Drug interactions between St. John’s wort and immunosuppressive drugs have been known for a long time. In patients who received organ transplantation, decreased blood concentration of cyclosporine was detected during the intake of St. John’s wort, and transplant graft rejection was observed. After stopping St. John’s wort, the blood concentration of cyclosporine was sharply increased and uncontrolled.214) On the other hand, down-regulation of CYP3A expression by drugs is considered to affect the pharmacokinetics of various drugs, such as DDI. However, to our knowledge, clinical DDIs through the downregulation of CYP3A have not been reported. Metformin, which was reported to cause CYP3A downregulation in vitro,140) is not known about clinical DDIs through the downregulation of CYP3A. DDI through inhibition of CYP3A enzymatic activity is also important, but is omitted in this review because it focuses on variations in CYP3A gene and protein expression.
The metabolic activation of drugs mediated by CYP3A can cause toxic effects. For example, acetaminophen overdose is known to stimulate metabolic activation by CYP3A4 into N-acetyl-p-benzoquinone imine (NAPQI), a major toxic metabolite, resulting in toxicity, such as liver failure, nephrotoxicity, and parkinsonism.215,216) CYP3A4 and CYP3A5 catalyze O-dealkylation of the dual-tyrosine kinase inhibitor lapatinib, which seems to induce idiosyncratic hepatotoxicity.217) Thalidomide treatment caused limb abnormalities in embryos of CYP3A humanized mice, but not in those of control mice, suggesting that human CYP3A7 in the placenta may mediate metabolic activation related to the teratogenicity of thalidomide.218) Thus, it is necessary to consider metabolic activation and decrease in drug blood concentration by CYP3A induction.
Table 4 shows drug-metabolizing enzymes related to the top 30 drugs reported in FDA Adverse Event Reporting System (FAERS) and Japanese Adverse Drug Event Report database (JADER). FAERS and JADER are databases containing adverse events-related reports that were submitted to FDA and PMDA, respectively. The top 30 most frequently reported drugs through the end of 2018 in FAERS analyzed by Kaneko and Nagashima were used.219) The top 30 most frequently reported drugs in JADER were analyzed using data through January 2021, and cases of the same drugs in the same patients were excluded for data cleaning (Table 4). Although the causal relationships between drug metabolizing-enzymes and adverse events are not proven, 11 in 30 products (FAERS) and 14 in 30 products (JADER) are partly metabolized by CYP3A (Table 4). Variation and inter-individual differences in CYP3A expression can contribute adverse effects of these products. In fact, the changes in pharmacokinetics and toxicity of several drugs in Table 4 such as acetaminophen, simvastatin, tacrolimus, cyclosporine, apixaban, and sorafenib, are shown in this review.
| FAERS | JADER | ||||
|---|---|---|---|---|---|
| Druga) | Drug metabolizing enzymes | Drug | Frequency | Drug metabolizing enzymes | |
| 1 | Adalimumab | —b) | Methotrexate | 22813 | AO |
| 2 | Etanercept | —b) | Prednisolone | 19812 | CYP3A4 |
| 3 | Aspirin | Esterase, Glycine N-acyltransferase, UGT | Tacrolimus | 12562 | CYP3A4, CYP3A5 |
| 4 | Thyroxine | Deiodinases, UGT, SULT, Deaminase, Decarboxylase | Bevacizumab | 11683 | —b) |
| 5 | Atorvastatin | CYP3A4 | Ribavirin | 10531 | —c) |
| 6 | Peritoneal dialysis solution | —d) | Nivolumab | 10017 | —b) |
| 7 | Acetaminophen | UGT, SULT, CYP2E1, CYP3A4, CYP1A2, GST | Cisplatin | 9884 | —d) |
| 8 | Interferon β-1a | —b) | Oxaliplatin | 9099 | —d) |
| 9 | Omeprazole | CYP2C19, CYP3A4 | Fluorouracil | 9055 | Dihydropyrimidine dehydrogenase |
| 10 | Furosemide | UGT | Acetylsalicylic acid | 8246 | Esterase, Glycine N-acyltransferase, UGT |
| 11 | Metformin | —d) | Cyclosporine | 7669 | CYP3A4 |
| 12 | Multi-vitamin | —c) | Tegafur/Gimeracil/Oteracil | 7373 | CYP2A6, Dihydropyrimidine dehydrogenase |
| 13 | Methotrexate | AO | Loxoprofen | 7364 | CYP3A4, CYP3A5, UGT2B7, Carbonyl reductase |
| 14 | Prednisone | CYP3A4 | Apixaban | 7305 | CYP3A4, CYP3A5 |
| 15 | Metoprolol | CYP2D6 | PEG Interferon α-2b | 6892 | —b) |
| 16 | Lenalidomide | —d) | Paclitaxel | 6880 | CYP2C8, CYP3A4 |
| 17 | Amlodipine | CYP3A4 | Carboplatin | 6836 | —d) |
| 18 | Natalizumab | —b) | Dexamethasone | 6646 | CYP3A4, CYP1A2, Carboxylesterase |
| 19 | Infliximab | —b) | Cyclophosphamide | 6416 | CYP2B6, CYP2C8, CYP2C9, CYP3A4, CYP2A6 |
| 20 | Simvastatin | CYP3A4 | Rivaroxaban | 6400 | CYP3A4, CYP3A5, CYP2J2 |
| 21 | Lisinopril | —d) | Irinotecan | 6393 | Carboxylesterase, UGT1A1, CYP3A4, CYP3A5 |
| 22 | Warfarin | CYP2C9, CYP1A2, CYP3A4, Carbonyl reductase | Lenalidomide | 6238 | —d) |
| 23 | Esomeprazole | CYP2C19, CYP3A4 | Pembrolizumab | 6200 | —b) |
| 24 | Quetiapine | CYP3A4, CYP2D6 | Carbamazepine | 6143 | CYP3A4 |
| 25 | Pregabalin | —d) | Docetaxel | 6013 | CYP3A4 |
| 26 | Salbutamol | SULT, UGT | Pregabalin | 5815 | —d) |
| 27 | Citalopram | CYP2D6, CYP2C19, CYP3A4, MAO, AO | Clopidogrel | 5807 | CYP2C19, CYP1A2, CYP2B6, CYP3A4 |
| 28 | Gabapentin | —d) | Sorafenib | 5341 | CYP3A4, UGT1A9 |
| 29 | Teriparatide | —c) | Tocilizumab | 5229 | —b) |
| 30 | Rivaroxaban | CYP3A4, CYP3A5, CYP2J2 | Gemcitabine | 5171 | Cytidine deaminase |
a): Analyzed by Kaneko and Nagashima,219) b): biological product (degradation into peptides or amino acids), c): unclear: d): no or little metabolized. UGT: uridine diphosphate glucuronosyltransferase, SULT: sulfotransferase, GST: glutathione S-transferase, AO: aldehyde oxidase, MAO: monoamine oxidase.
CYP3A expression is altered by various endogenous and exogenous factors, as described above. Various factors collaboratively cause inter- and intra-individual differences in CYP3A expression and activity. The variation in CYP3A expression is clinically relevant to pharmacokinetics during pharmacotherapy. In addition, it can contribute to disease development and progression and may be a predictor of the disease. In addition, CYP3A contributes to the maintenance of homeostasis of endogenous substances, and its disruption might be associated with toxicity. Therefore, factors affecting CYP3A expression and the toxicological effects caused by changes in CYP3A expression must be clarified further. An accurate prediction of individual differences in CYP3A function is expected in the future.
We would like to thank Dr. Keiko Ogawa of Ritsumeikan University for analyzing adverse drug event reports in JADER.
The authors declare no conflict of interest.