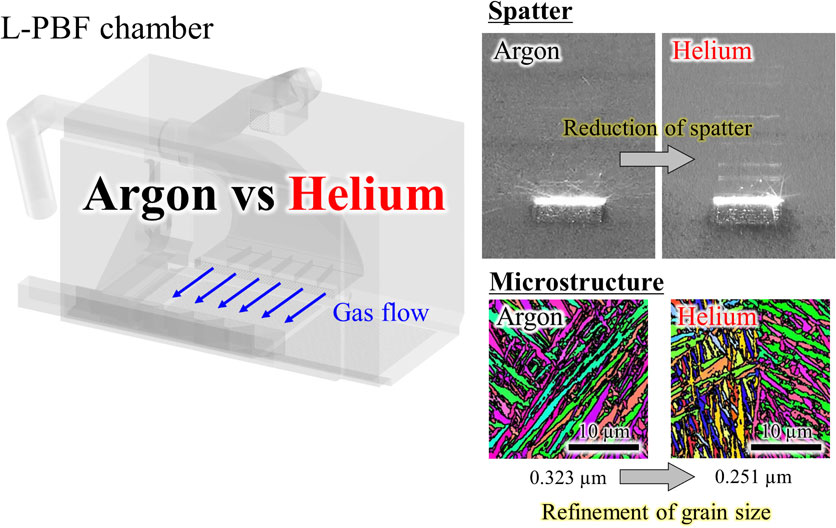
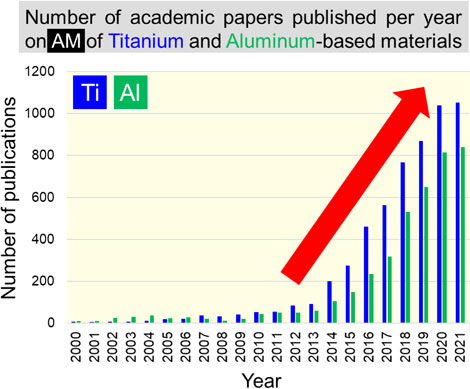
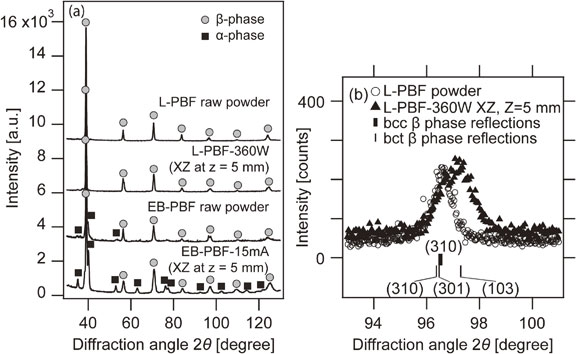
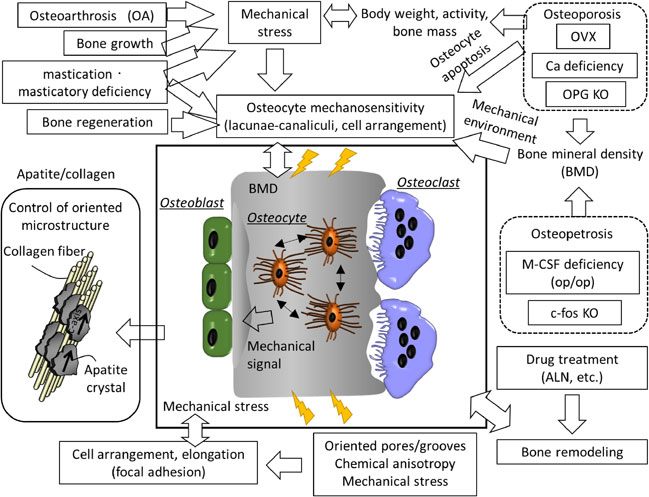
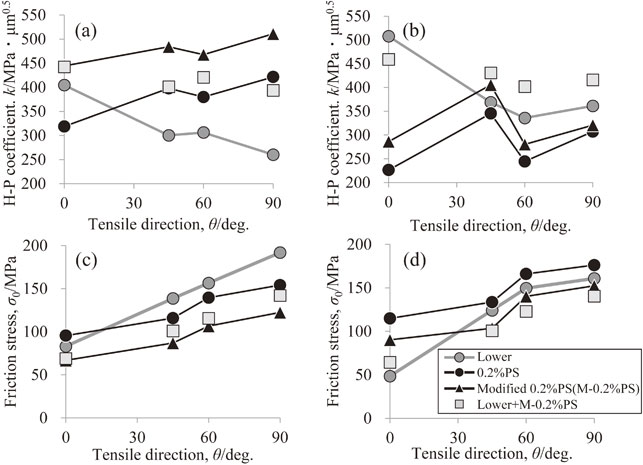
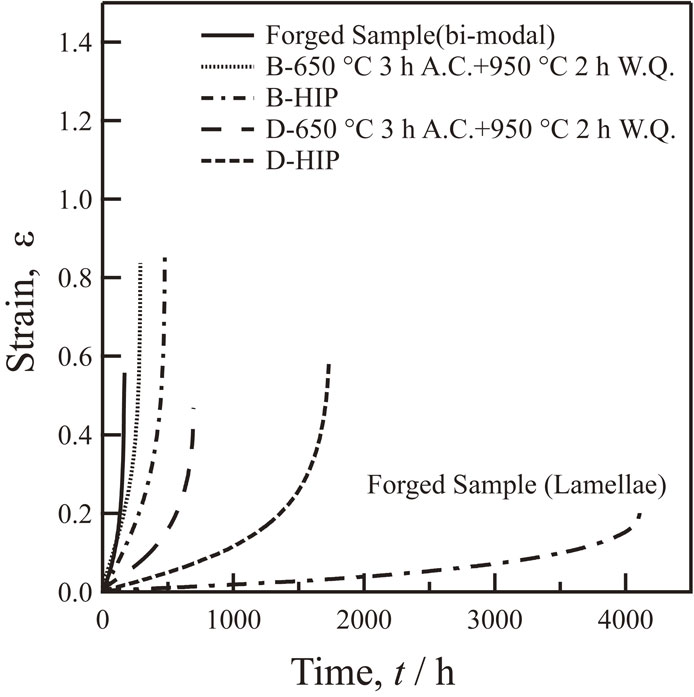
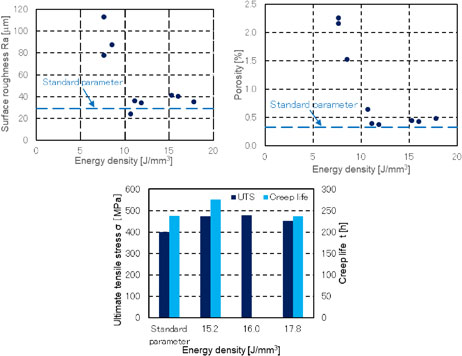
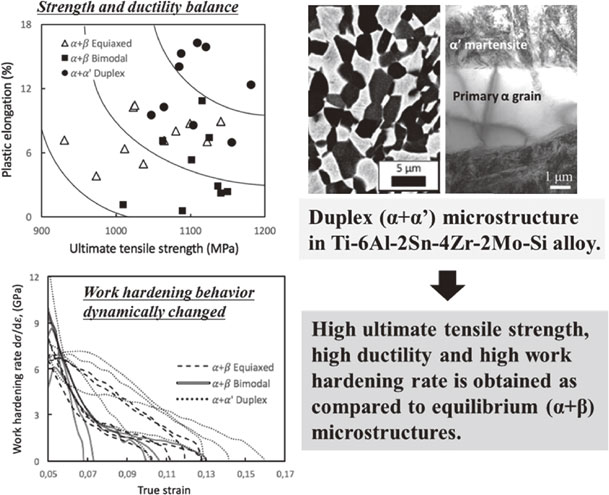
Volume 64, Issue 1
Displaying 1-42 of 42 articles from this issue
- |<
- <
- 1
- >
- >|
Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
-
Article type: Preface
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 1
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (28K) Full view HTML -
Article type: Review
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 2-9
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: September 16, 2022Download PDF (5494K) Full view HTML -
Article type: Review
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 10-16
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 17, 2022Download PDF (4669K) Full view HTML -
Article type: Review
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 17-24
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: August 26, 2022Download PDF (4903K) Full view HTML -
Article type: Review
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 25-30
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 14, 2022Download PDF (3183K) Full view HTML -
Article type: Review
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 31-36
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 21, 2022Download PDF (4507K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 37-43
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 21, 2022Download PDF (4792K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 44-49
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (2553K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 50-53
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 17, 2022Download PDF (1130K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 54-60
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: December 05, 2022Download PDF (5856K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 61-70
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (2288K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 71-77
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: August 26, 2022Download PDF (4553K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 78-85
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (1050K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 86-94
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: September 30, 2022Download PDF (1723K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 95-103
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 28, 2022Download PDF (5222K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 104-110
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (4608K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 111-120
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (4901K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 121-130
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 17, 2022Download PDF (9393K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 131-137
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: September 30, 2022Download PDF (2215K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 138-146
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: December 05, 2022Download PDF (3101K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 147-154
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 17, 2022Download PDF (3348K) Full view HTML -
Article type: Regular Article
Subject area: Special Issue on Recent Research and Development in the Processing, Microstructure, and Properties of Titanium and Its Alloys
2023Volume 64Issue 1 Pages 155-164
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: October 21, 2022Download PDF (3540K) Full view HTML
Regular Article
Materials Physics
-
Article type: Regular Article
2023Volume 64Issue 1 Pages 165-170
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 28, 2022Download PDF (2315K) Full view HTML
Microstructure of Materials
-
Article type: Regular Article
2023Volume 64Issue 1 Pages 171-176
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (3800K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 177-183
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 28, 2022Download PDF (4986K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 184-190
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (1900K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 191-195
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (3692K) Full view HTML
Mechanics of Materials
-
Article type: Regular Article
2023Volume 64Issue 1 Pages 196-204
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 04, 2022Download PDF (2989K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 205-211
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 14, 2022Download PDF (3393K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 212-219
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 14, 2022Download PDF (9284K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 220-226
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (4581K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 227-232
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (6381K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 233-241
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 28, 2022Download PDF (3561K) Full view HTML
Materials Chemistry
-
Article type: Regular Article
2023Volume 64Issue 1 Pages 242-251
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (4767K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 252-259
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 28, 2022Download PDF (5655K) Full view HTML
Materials Processing
-
Article type: Regular Article
2023Volume 64Issue 1 Pages 260-266
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: November 18, 2022Download PDF (3080K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 267-271
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: December 05, 2022Download PDF (1273K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 272-279
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (3438K) Full view HTML
Engineering Materials and Their Applications
-
Article type: Regular Article
2023Volume 64Issue 1 Pages 280-286
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (1744K) Full view HTML -
Article type: Regular Article
2023Volume 64Issue 1 Pages 287-295
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (3306K) Full view HTML
Technical Article
-
Article type: Technical Article
2023Volume 64Issue 1 Pages 296-302
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Advance online publication: December 05, 2022Download PDF (4833K) Full view HTML
Express Rapid Publication
-
Article type: Express Rapid Publication
2023Volume 64Issue 1 Pages 303-305
Published: January 01, 2023
Released on J-STAGE: December 25, 2022
Download PDF (1686K) Full view HTML
- |<
- <
- 1
- >
- >|