- |<
- <
- 1
- >
- >|
-
Yutaka KURODA2022Volume 62Issue 5 Pages 267
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (144K) Full view HTML
-
Keisuke SHIMIZU, Masataka USAMI, Ikuro MIZOGUCHI, Shoko FUJITA, Ryuji ...2022Volume 62Issue 5 Pages 271-275
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDe novo protein design has emerged as a method to manipulate the primary structure for the development of artificial proteins and peptides with desired functionality. This paper describes the de novo design of a pore-forming peptide that has a β-hairpin structure and assembles to form a stable nanopore in a bilayer lipid membrane. We designed two kinds of peptides, SV28: forming multidispersely-sized nanopore and SVG28: monodispersely-sized nanopore, and succeeded to detect single molecule DNAs and polypeptides. Such de novo design of a β-hairpin peptide has the potential to create artificial nanopores, which can be adjust size to a target molecule.
 View full abstractDownload PDF (1409K) Full view HTML
View full abstractDownload PDF (1409K) Full view HTML -
Keita KAMINO2022Volume 62Issue 5 Pages 276-279
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLOrganisms acquire and use sensory information to guide their behaviors. However, it is unclear whether and how this information constrains the ability of organisms to perform behavioral tasks. In a recent work, using E. coli chemotaxis as a model system, we showed that the sensory information that a bacterium acquires sets an upper limit on its behavioral performance. Furthermore, combined with quantitative experiments, we quantified the rate at which E. coli acquire information during navigation and discovered that E. coli use the acquired information efficiently. Here, we succinctly review the key findings of the work.
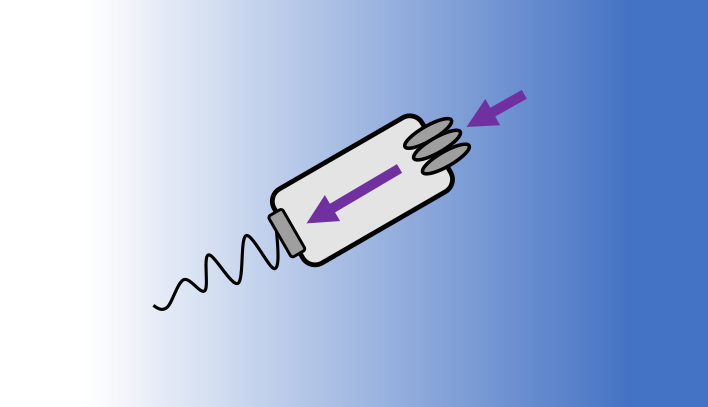 View full abstractDownload PDF (822K) Full view HTML
View full abstractDownload PDF (822K) Full view HTML -
Yuki HARA2022Volume 62Issue 5 Pages 280-284
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLThe eukaryotic cells can change the size of the cell nucleus, including the genomic DNA, depending on the environment surrounding the cell nucleus. How cells can scale the nuclear size by sensing the environment has remained an enigma for over a century in the field of cell biology. Here, we found a general scaling rule of nuclear size to cell size among eukaryotes and another new aspect for size scaling to the chromatin status inside the nucleus. In this review, I describe the known mechanisms underlying these size scaling characteristics and discuss putative physiological significances for the size scaling.
 View full abstractDownload PDF (1381K) Full view HTML
View full abstractDownload PDF (1381K) Full view HTML
-
Kei YAMAMOTO, Yohei KONDO2022Volume 62Issue 5 Pages 285-287
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (1887K) Full view HTML -
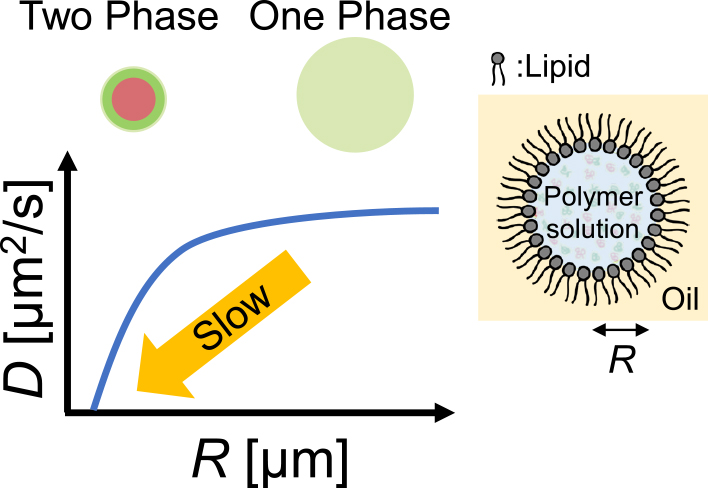
Akira NAGASAKI, Taro Q. P. UYEDA2022Volume 62Issue 5 Pages 288-290
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (1653K) Full view HTML -
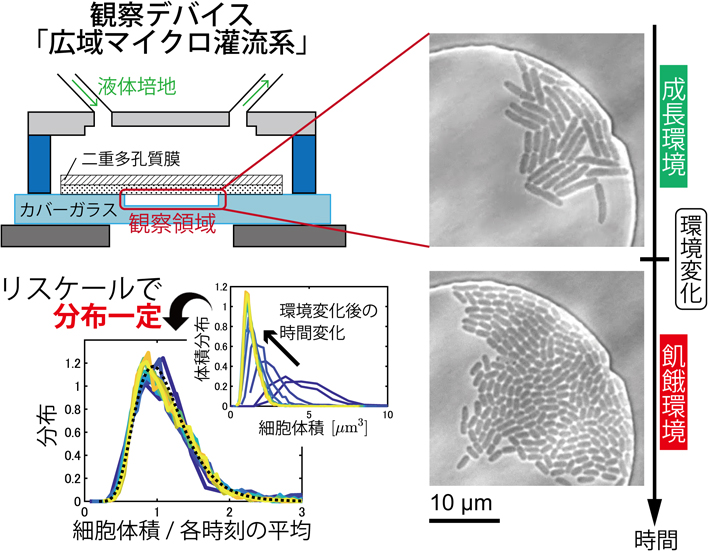
Kazumasa A. TAKEUCHI, Takuro SHIMAYA2022Volume 62Issue 5 Pages 291-294
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (2511K) Full view HTML -
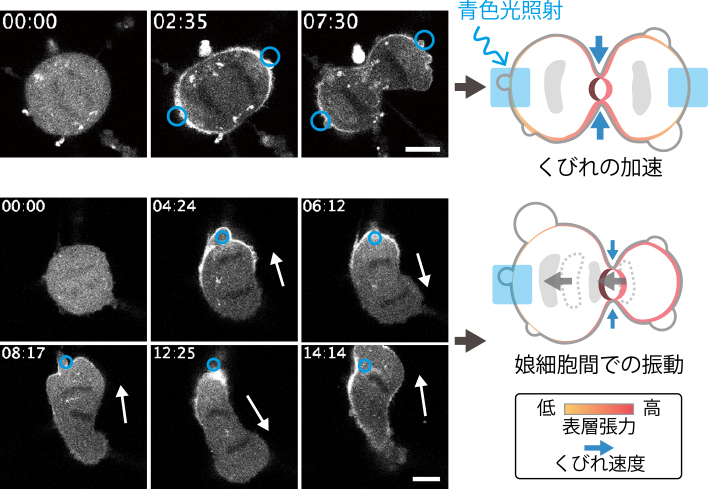
Chika OKIMURA, Yoshiaki IWADATE2022Volume 62Issue 5 Pages 295-297
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (1046K) Full view HTML -
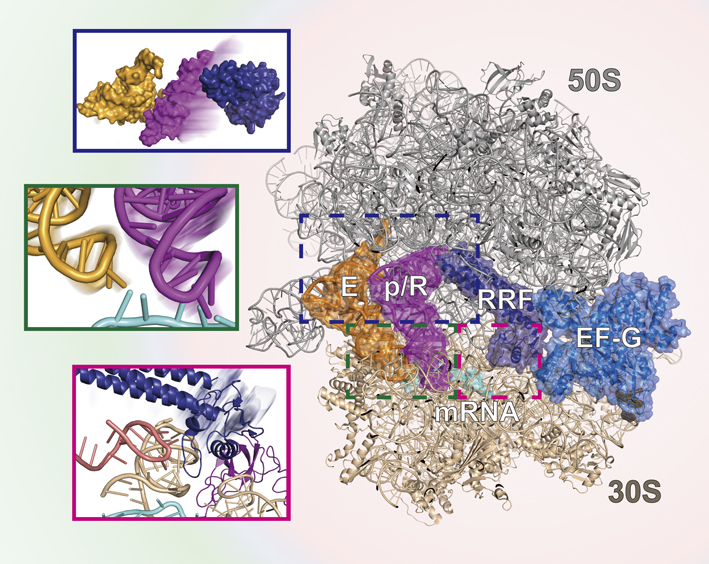
Chigusa KOBAYASHI, Yasuhiro MATSUNAGA, Jaewoon JUNG, Yuji SUGITA2022Volume 62Issue 5 Pages 298-300
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (2073K) Full view HTML
-
Chiho WATANABE2022Volume 62Issue 5 Pages 301-302
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (1225K) Full view HTML -
Takehito TANZAWA2022Volume 62Issue 5 Pages 303-304
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (2039K) Full view HTML
-
Hiroyuki MINO2022Volume 62Issue 5 Pages 307-309
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (1606K) Full view HTML -
Shinya HONDA2022Volume 62Issue 5 Pages 310-311
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (653K) Full view HTML
-
[in Japanese]2022Volume 62Issue 5 Pages 312-313
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (2008K) Full view HTML
-
[in Japanese]2022Volume 62Issue 5 Pages 314-315
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (1041K) Full view HTML
-
[in Japanese]2022Volume 62Issue 5 Pages 316-317
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (965K) Full view HTML
-
2022Volume 62Issue 5 Pages 305-306
Published: 2022
Released on J-STAGE: November 25, 2022
JOURNAL FREE ACCESS FULL-TEXT HTMLDownload PDF (165K) Full view HTML
- |<
- <
- 1
- >
- >|